Structural Biology
Hydrogen Deuerium exchange mass spectrometry (HDX MS)
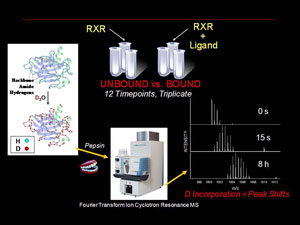
Our structural biology efforts have focused on the use of Hydrogen deuterium exchange (HD X) coupled with high resolution mass spectrometry (MS) to analyze changes in protein dynamics upon binding of other proteins or ligands. We have a dedicated LTQ-FT MS instrument (HeiDi) coupled with a liquid handling robot/HPLC interface (LEAP Technologies)

HeiDi
Our HDX MS efforts at UAB have been lead by Dr. Peter Prevelige in the Department of Microbiology. Dr. Prevelige was one of the early adopters of HDX MS methodologies in the analysis of viral structure and assembly. More information on Dr. Prevelige’s work can be found here.
- Cortines JR, Monroe EB, Kang S, Prevelige PE Jr. A retroviral chimeric capsid
protein reveals the role of the N-terminal β-hairpin in mature core assembly. J
Mol Biol. 2011 Jul 22;410(4):641-52. doi: 10.1016/j.jmb.2011.03.052. PubMed PMID:
21762805; PubMed Central PMCID: PMC3139139.
- Monroe EB, Kang S, Kyere SK, Li R, Prevelige PE Jr. Hydrogen/deuterium
exchange analysis of HIV-1 capsid assembly and maturation. Structure. 2010 Nov
10;18(11):1483-91. doi: 10.1016/j.str.2010.08.016. PubMed PMID: 21070947; PubMed
Central PMCID: PMC2982214.
- Kang S, Poliakov A, Sexton J, Renfrow MB, Prevelige PE Jr. Probing conserved helical modules of portal complexes by mass spectrometry-based hydrogen/deuterium exchange. J Mol Biol. 2008 Sep 5;381(3):772-84. doi: 10.1016/j.jmb.2008.03.004. Epub 2008 Mar 18. PubMed PMID: 18621389; PubMed Central PMCID: PMC2593103.
Renfrow Laboratory use of HDX MS
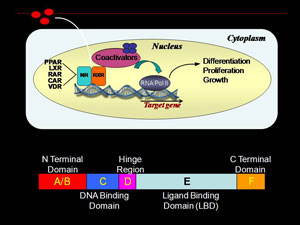
Retinoid X receptor (RXR) is a ligand dependent nuclear receptor transcription factor fundamental in regulation of cellular proliferation, differentiation and growth. Small molecule binding repositions key structural elements in RXR ligand binding domain (LBD), presenting/burying coregulator interaction surfaces. Agonist binding removes corepressor proteins and promotes interactions with transcriptional coactivators. RXR associated transcription complexes are of therapeutic interest for prevention and treatment of breast cancer. We are interested in the process of how ligand binding results in a conformational change in the ligand binding domain of RXR nuclear receptor transcription factor that leads to a specific transcriptional response. This ligand dependent process is often studied by use of X-ray crystallography. However, many of these resulting structures are very similar.
The Renfrow laboratory makes use of HDX MS compare and contrast the changes in in-solution conformational dynamics of the RXR LBD when bound to different ligands and coactivators. Our current work centers around the UAB developed 9-cis-UAB30, an RXR specific retinoid that has shown great promise a chemopreventative agent. These HD X profiles provide a dynamic component to the structural analysis and can detect differences that ligand binding induces in the RXR LBD that are not observed in x-ray crystal analysis. These dynamic changes are quantative and can be used as structural markers to screen novel rexinoids for similar binding characteristics. We also correlate our results with more traditional molecular biology assays for characterizing novel RXR ligands in terms of ligand affinity and RXR activation. Overall, our goal is to provide a unique set of tools and perspective for understanding the process of ligand induced nuclear receptor activation.
- Boerma LJ, Xia G, Qui C, Cox BD, Chalmers MJ, Smith CD, Lobo-Ruppert S, Griffin P, Muccio DD, Renfrow MB.
Defining the communication between agonist and coactivator binding in the retinoid X receptor ligand binding domain.
J Biol Chem. 2014 Jan 10;289(2):814-26.
- Xia G, Boerma LJ, Cox BD, Qiu C, Kang S, Smith CD, Renfrow MB, Muccio DD.
Structure, Energetics, and Dynamics of Binding Coactivator Peptide to the Human Retinoid X Receptor α Ligand Binding Domain Complex with 9-cis-Retinoic Acid.
Biochemistry. 2011 Jan 11;50(1):93-105. Epub 2010 Dec 8.
Research Support
This work has been supported by: