New Techniques Could Accelerate Drug Discovery
By Matt Windsor
 In his UAB lab, Stephen Aller (far left) is exploring new ways to dramatically speed up the drug-discovery process for cystic fibrosis, cancer, and other diseases. Aller's research team includes (left to right) Zachary Tibbs (standing), Kimberly Jaimes (sitting), Lindsay Turner, Jessica Makofske, and Jingzhi Li.
In his UAB lab, Stephen Aller (far left) is exploring new ways to dramatically speed up the drug-discovery process for cystic fibrosis, cancer, and other diseases. Aller's research team includes (left to right) Zachary Tibbs (standing), Kimberly Jaimes (sitting), Lindsay Turner, Jessica Makofske, and Jingzhi Li.
If you could look inside the lungs of a child with cystic fibrosis, you would see a layer of thick, sticky gunk coating every surface, providing a rich haven for bacteria and making it very hard to breathe. If you could look into the child’s future, the view wouldn’t be pretty, either. Every day, her parents will have to pound on her back in order to break up the accumulated mucus and allow her to cough it out. While previous generations of patients with cystic fibrosis often died in their teens, this child will at least have a good chance of living into her 30s. However, her lung disease will have worsened to the point that she will become disabled and, eventually, die.
Medications have played a major role in extending the lifespan of patients with cystic fibrosis. Antibiotics help prevent serious lung and sinus infections, inhaled medicines open the airways, and enzyme therapies can thin mucus. But the root of the problem, revealed in 1989 in part by UAB scientists, lies in a single defective gene. And at the moment doctors don’t have anything that can touch it.
The gene produces a protein called the cystic fibrosis transmembrane conductance regulator (CFTR). When CFTR is functioning normally, it shuttles chloride and thiocyanate ions through cell membranes, which helps keep mucus thin and flowing freely. But when CFTR is defective, the channels can become partially or completely blocked, or they may never become embedded in the cell wall in the first place.
Getting Through
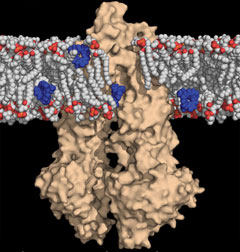 In this image, cancer drugs (in blue) intended to penetrate the membrane boundary of a cancer cell (white and red) are intercepted by a molecular drug pump called P-glycoprotein (in tan) and expelled out of the cell, resulting in multi-drug resistant tumors in the body.If scientists could find a way to restore normal function to the CFTR channels, they could reduce or even reverse the effects of cystic fibrosis. Some promising treatments in the pipeline do just that—including one drug, VX-770, currently in advanced clinical trials at UAB. Despite these successes, however, scientists are laboring in the dark. They don’t know exactly how CFTR works, and that means progress must come as a result of trial and error, says Stephen G. Aller, Ph.D., an assistant professor in the UAB Department of Pharmacology and Toxicology and associate scientist in the Experimental Therapeutics program at the UAB Comprehensive Cancer Center.
In this image, cancer drugs (in blue) intended to penetrate the membrane boundary of a cancer cell (white and red) are intercepted by a molecular drug pump called P-glycoprotein (in tan) and expelled out of the cell, resulting in multi-drug resistant tumors in the body.If scientists could find a way to restore normal function to the CFTR channels, they could reduce or even reverse the effects of cystic fibrosis. Some promising treatments in the pipeline do just that—including one drug, VX-770, currently in advanced clinical trials at UAB. Despite these successes, however, scientists are laboring in the dark. They don’t know exactly how CFTR works, and that means progress must come as a result of trial and error, says Stephen G. Aller, Ph.D., an assistant professor in the UAB Department of Pharmacology and Toxicology and associate scientist in the Experimental Therapeutics program at the UAB Comprehensive Cancer Center.
In a process known as high-throughput screening, Aller explains, pharmaceutical companies use advanced robotics and computer hardware to test thousands of different compounds, and then focus on the most promising candidates. The technology is impressive, but it’s also expensive and time-consuming, Aller notes. “It can take years and cost millions of dollars.” Aller is looking for a better way. He is a structural biologist—which means he studies the underlying molecular architecture of proteins and other large molecules in order to understand their function—and he believes that structure-based drug design has the potential to dramatically speed up the drug pipeline for cystic fibrosis and many other diseases.
|
Pharmaceutical companies use advanced robotics and computer hardware to seek out promising new drugs. But that process "can take years and cost millions of dollars," Aller says. He is searching for a better way. |
In his UAB lab, Aller is developing novel methods to take high-resolution snapshots of CFTR and other similar proteins that play a role in disease. With this information in hand, scientists could figure out precisely how these proteins function and have a much better idea of how to develop new drugs to target them.
While he was at the Scripps Research Institute in California, Aller made a breakthrough in the field, obtaining the first structural “solution” for an important drug transporter protein known as P-glycoprotein. (“Solving” a protein means you have determined its underlying three-dimensional molecular structure.) In September 2011, the National Institutes of Health (NIH) recognized the potential of Aller’s work with a five-year, $1.5-million Director’s New Innovator Award. The prestigious honors aim to stimulate highly innovative research and support promising new investigators.
Sentry Duty
Aller’s lab is focusing its attention on human membrane proteins, which reside in the double-layered lipid fence that surrounds our cells. The proteins function as gatekeepers; their job is to allow approved substances in and pump other substances out. Membrane proteins play a vital role in maintaining the blood-brain barrier, for example, and they are also crucial to the filtering functions of the kidneys, liver, and intestines. “A third of all the drugs that are approved by the FDA and now on the market target this class of proteins,” Aller says. “And the membrane proteins are involved in many diseases, including diabetes, multi-drug resistance in cancer, and AIDS-related dementia, in addition to cystic fibrosis.”
Chemical WarfareHow Membrane Proteins “Solving” the structures of human membrane proteins could open up a new front in the fight against cancer, says UAB structural biologist Stephen G. Aller, Ph.D. “Most of the time, if you’re giving a chemotherapeutic agent, it will probably kill the cancer the first time around,” Aller says. “The patient goes home well, but sometimes the cancer comes back. Then the doctors will try to kill it again using the same therapy, and it often doesn’t work: The cancer cell has figured out how to become resistant.” This drug resistance often can be traced back to membrane proteins, Aller explains. “Cancer cells start to overproduce transporters that can pump out the drugs.” That’s actually doubly bad news, Aller says, because the transporters are “polyspecific,” which means they recognize thousands of different compounds. “Even if you switch to a chemically unrelated therapeutic agent, it may not work either.” Most cancers have the capability of becoming drug resistant, Aller says, but figuring out how to disrupt their transporter proteins could give scientists a way to fight back. “The cancer cell turns up its defense mechanisms to protect itself against the chemotherapeutic agents,” he says. “We have to respond.” |
Some membrane proteins, like CFTR, act as channels—when they are open, they let in a flood of chemical substrates; when they close, nothing gets through. But most membrane proteins are of a type known as transporters. “They expose a binding site to the outside of the cell; when they capture their target molecule, they pivot around it and expose their binding site to the inside, or vice versa,” Aller says. “Then the molecule is shuttled across the membrane, and the transporter resets itself for another cycle.” The same proteins may transport a variety of different substances, including neurotransmitters the body produces and chemicals introduced from the outside, such as the components in a dose of aspirin. There may be hundreds of individual proteins of each type on an individual cell membrane, Aller notes. “It’s very densely packed.”
Membrane proteins are “molecular machines,” Aller continues. They move through various configurations as they open to expel unwanted material from a cell and then close to maintain the cell’s integrity. Aller’s goal is to use X-ray crystallography to take a series of snapshots of human membrane proteins as they cycle through these configurations. “An X-ray beam is very thin,” he explains. “When you shoot it at a crystal, it diffracts off all the atoms inside and gives you a pattern. By turning the crystal, you get different patterns, and then you can use a computer to figure out what the object was that produced that pattern.”
With detailed computer models of membrane proteins in hand, researchers can identify the precise areas they need to target—and have a much stronger idea of what drugs will be most effective. This is known as structure-based drug design.
New Approach
Structure-based drug design, according to Aller’s rough calculations, could speed up the process of finding new treatments by at least a year and save millions of dollars in development costs. But to practice structure-based design, scientists need precise information about the various conformations of human membrane proteins. And those efforts have run into several roadblocks. One is synthesizing enough samples of each target protein to produce the crystals researchers require for X-ray crystallography. “Another major obstacle is that some of the proteins intrinsically don’t behave well,” Aller says. “They like to stick together, for example, which is bad for the crystallization process.”
|
To practice structure-based drug design, scientists need precise information about the various conformations of human membrane proteins. Aller's lab is developing new ways to capture high-resolution snapshots of these proteins in action. |
Overcoming these roadblocks is what the NIH Innovator award is all about, Aller explains. “We’re really thinking outside the box in terms of how to accelerate the production and the crystallization of membrane proteins.” Strategies include using molecular “chaperones” to encourage the target proteins to fold properly, synthetic scaffolds to hold the proteins in particular conformations, and model systems such as yeast in order to speed protein growth.
UAB’s existing strengths in cystic fibrosis research helped attract Aller to Birmingham in 2009. His team is now “aggressively pursuing” the structure of the CFTR protein. “A couple of pretty good drugs are being tested now, but because we don’t know the structure of that protein, we don’t know why they’re working,” Aller says. “Are they directly interacting with the protein, or are they doing something ancillary? That’s an easy hypothesis to test if you have the structure modeled in a computer. The quickest thing you get from a structure is the ability to generate quick and relatively accurate hypotheses that give you directions for further research. We could say, ‘This one works, and that one works in a different way, so why don’t we try the two together because they might synergize?’”
If scientists can identify 20 compounds with potential to be effective treatments for a disease, “and we can start to classify them based on their interactions with a protein’s structure, then we can test them in pairs. That can save time by orders of magnitude.”
Although structure-based drug design is still in its infancy, there are already some success stories, including drugs to treat HIV and glaucoma, Aller notes. “Our own work is in the early stages, but already we’re getting pretty good hits in cystic fibrosis cells,” he says. “We’re really excited about the potential.”