Need more information? Contact us
 Train a computer to read medical records, and you could do a world of good. Doctors could use it to look for dangerous trends in their patients’ health. Researchers could speed drugs to market by quickly finding appropriate patients for clinical trials. They could also find previously overlooked associations. By keeping track of data points across tens of thousands, or millions, of medical records, computer models could find patterns that would never occur to individual researchers. Maybe Asian women in their 40s with type 2 diabetes respond well to a certain combination of medications, while white men in their 60s do not, for example.
Train a computer to read medical records, and you could do a world of good. Doctors could use it to look for dangerous trends in their patients’ health. Researchers could speed drugs to market by quickly finding appropriate patients for clinical trials. They could also find previously overlooked associations. By keeping track of data points across tens of thousands, or millions, of medical records, computer models could find patterns that would never occur to individual researchers. Maybe Asian women in their 40s with type 2 diabetes respond well to a certain combination of medications, while white men in their 60s do not, for example.
Machine learning, in particular a branch called natural language processing, has had plenty of successes recently. It’s the secret sauce behind IBM’s “Jeopardy”-winning Watson computer and Apple’s Siri personal assistant, for instance. But computers still have a tough time following medical narratives.
“We take it for granted how easy it is for us to understand language,” said Steven Bethard, Ph.D., a machine learning expert and linguist in the UAB College of Arts and Sciences Department of Computer and Information Sciences. “When I’m having a conversation, I can use all kinds of crazy constructions and pauses between words, and you would still understand me. All these things make language very difficult for computers, however. They like rules and an order that is followed every time, but languages aren’t like that.”
Timing is everything
So Bethard, the director of UAB’s Computational Representation and Analysis of Language Lab, builds models that help computers catch our drift. In one ongoing project, he is working with colleagues at the Mayo Clinic and Boston Children’s Hospital “to extract timelines from clinical work,” Bethard said. Using text from clinical notes taken at the Mayo Clinic, “we’re working to find all the clinical events mentioned in those notes — things like ‘asthma’ and ‘CT scan,’ for example — and link them to the proper time,” he said. If the computer sees “the patient has a history of asthma,” it should know that’s in the past. If it sees “planning a CT scan,” that’s in the future. “Sometimes you have explicit dates, such as ‘on Sept. 15, the patient had a colonoscopy,’” Bethard said. “But the computer still has to figure out whether that means Sept. 15, 2014, or Sept. 15, 2015.’”
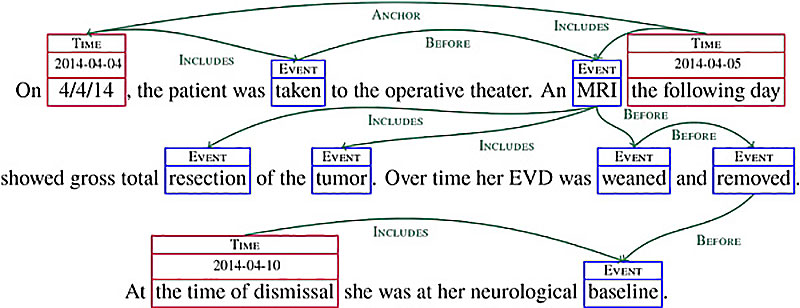 The diagram above illustrates how a computer could extract timeline information out of an entry in a medical record.A system like this would help individual doctors keep track of their patients’ progress. “If you have had a patient for 15 years, you see so many things,” Bethard said. “Looking at a visual of all the conditions and procedures over that time is extremely useful.” The system could also identify patients for clinical trials. “Say you wanted to find someone who had liver toxicity after they started taking methotrexate,” Bethard said. “The sequence of events is important; you only want to find people who have taken the drug and had liver toxicity in the appropriate order.” Another use: finding new associations between drugs or procedures and adverse events. “If you have a large number of patients, you can say, ‘How often do you see a certain side effect?’ for example,” Bethard said. “You can generate new hypotheses about causality.”
The diagram above illustrates how a computer could extract timeline information out of an entry in a medical record.A system like this would help individual doctors keep track of their patients’ progress. “If you have had a patient for 15 years, you see so many things,” Bethard said. “Looking at a visual of all the conditions and procedures over that time is extremely useful.” The system could also identify patients for clinical trials. “Say you wanted to find someone who had liver toxicity after they started taking methotrexate,” Bethard said. “The sequence of events is important; you only want to find people who have taken the drug and had liver toxicity in the appropriate order.” Another use: finding new associations between drugs or procedures and adverse events. “If you have a large number of patients, you can say, ‘How often do you see a certain side effect?’ for example,” Bethard said. “You can generate new hypotheses about causality.”
Learning to spot cancer
One of Bethard’s graduate students, John David Osborne, has built a machine-learning model that is already having an impact on the practice of health care at UAB. By day, Osborne is a research associate in the biomedical informatics group of UAB’s Center for Clinical and Translational Science. He and his colleagues were called in to help UAB’s Cancer Registry with a Big Data challenge: tracking and cataloguing cancer diagnoses and treatment outcomes.
Every hospital is responsible for reporting new cases of many different types of diseases to the federal government. “Cancer is one of those diseases, but not all cancers are reportable,” Osborne said. “Lots of skin cancers aren’t, but melanoma is; anything malignant or in the central nervous system is reportable.” Identifying and tracking these cases in pathology reports — and determining whether they are or are not reportable — can be quite challenging at a health care system as large as UAB, Osborne notes. A year and a half ago, the biomedical informatics team at the CCTS created the Cancer Registry Control Panel, which uses natural language processing to detect possible cancer cases in the pathology reports. As an additional research project, Osborne recently designed a machine-learning algorithm that provides additional assistance to the human registrars. “It scans through the records and says, ‘This is a likely case, and here’s why I think that,’” Osborne said. “Humans are still going through every record, but you can speed it up and show them where to look.”
 Language matters
Language matters
Bethard and Osborne build their models using the Unstructured Information Management Architecture — an open-source version of the code IBM used to create Watson.
The first step in building a machine-learning model is to decide what kind of training material to use. “The machine-learning models we create for health information extraction look at gold-standard models that humans have created,” Bethard said. “They say, ‘I see all these patterns in the human timelines, so this is what I’ll look for.’”
Some of these decisions are relatively simple. “Cancer is always a condition of interest,” Bethard said. “Anything related to cancer is something you want to include. The harder pattern to learn is how to link together time and events. A date and then a colon tells you they are describing something that happened on that date. Verb phrases, noun phrases and linguistic structure in time can be very predictive.”
As that description makes clear, natural language processing requires a deep knowledge of English grammar as well as computer code. “The most successful people in this field are hybrids,” adept at linguistics and computer science, Bethard said. He has a bachelor’s degree in linguistics. He shares his interest in language with his wife, who is now completing a postdoctoral fellowship in the cognitive neuroscience of language at the University of South Carolina.
Bethard came to Birmingham in 2013, attracted by ongoing research in natural language processing in UAB’s computer science department. “For me, it makes a lot of sense to be at a place with a major medical school,” Bethard said. He is looking forward to collaborations with James Cimino, M.D., Ph.D., the inaugural director of the School of Medicine’s new Informatics Institute and a renowned expert in the creation and manipulation of electronic medical records. “He’s famous, very well-known and well-respected,” Bethard said of Cimino. “He knows about all the range of problems: getting information from the text that doctors write, how to input this data, how to store it — the whole spectrum.”
Teaching computers to navigate the ambiguity of the English language can be trying, but the opportunities at UAB are exciting, Bethard says. “There is plenty of data available here, and clear challenges for these models to address.”