Author: Daniel Gilliam
The remarkable ability of cells in the brain to store and process information has fascinated scientists for centuries. To accomplish this feat, neurons must have mechanisms to generate both transient and persistent changes in response to incoming information. One mechanism proposed to mediate both long and short term changes in neurons is epigenetics.
Cells use epigenetics to modify and regulate their genome without altering the underlying DNA sequence. The prefix “epi,” meaning above, illustrates that epigenetic processes do not alter the information contained in DNA. Instead, they alter how and whether this information is used, often by regulating the three-dimensional structure of chromatin and the ability of proteins to bind at specific sites. Traditionally, epigenetic modifications were considered permanent, heritable markers that determine the sets of genes active within a given cell and its progeny. Because coordinated programs of gene expression underlie differentiation, epigenetics is known to play a crucial role in development. However, in the past two decades, investigators have discovered that the molecular machinery of epigenetics has been co-opted by the nervous system to mediate both dynamic and persistent changes in gene expression that underlie learning and memory, a process sometimes referred to as neuroepigenetics1.
Two main epigenetic mechanisms have been implicated in learning and memory: DNA methylation and histone modification2,3. DNA methylation involves the addition of a methyl group (-CH3) to cytosine residues within DNA. Typically, only cytosine residues adjacent to guanine are methylated, and such pairs are identified as “CpG” sequences (the “p” refers to the phosphate group in the intervening covalent bond). The distribution of these CpG pairs in the genome is much lower than would be predicted according to random chance. Although approximately 70% of these pairs are methylated, those in regions involved in regulating transcription are largely unmethylated. DNA methylation usually represses transcription, especially when it occurs within the promoter region. This repression is attributed to changes in interactions between proteins and DNA; some proteins have methyl binding domains and will only recognize a sequence if it is methylated1.
A crucial discovery implicating epigenetics in learning and memory was the observation that neuronal activity can induce changes in DNA methylation4. As one neuron communicates with another, signaling cascades are activated in the receiving neuron whichcan ultimately lead to alterations in the epigenetic status of particular genes. Because many different genes can be loosely defined as memory-permitting or memory-suppressing, transient changes in gene methylation provide a potential mechanism for temporary recruitment or suppression of genes to facilitate memory formation. For example, in the mouse hippocampus during fear learning the memory promoting gene Reelinis rapidly demethylated and transcriptionally activated. Similarly, the memory suppressing gene PP1is rapidly methylated and deactivated4.
A few hours after fear conditioning, the methylation status of these genes in hippocampal neurons returns to its original level4. Such observations suggest the existence of both active methylation and demethylation mechanisms. DNA methylation is known to be catalyzed by DNA methyltransferases (DNMTs), but the mechanisms of demethylation are less clear. Some have proposed that demethylation takes advantage of mechanisms that otherwise function in cellsto repair DNA base pair mismatches1. Figure 1B shows a proposed mechanism for demethylation of DNA by such a pathway. Some studies have suggested that intermediates in this pathway do occur in the central nervous system and may serve as precursors for DNA demethylation5,6.
Whatever the mechanisms, it seems that active demethylation can play a role in a neuron’s return to basal gene expression after a learning event4.
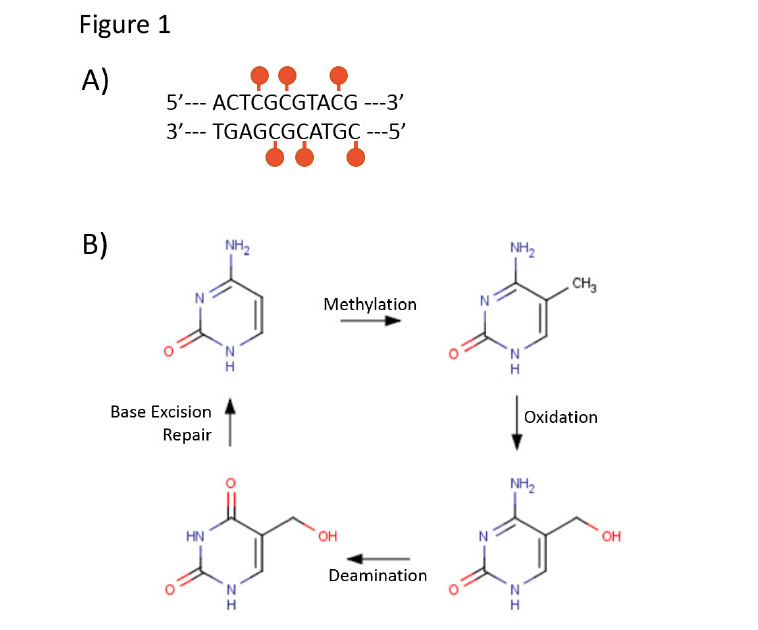
Figure 1 | Proposed mechanism of DNA methylation and demethylation. A) Within the genome, the majority of CpG pairs are methylated by addition of –CH3 to cytosine. B) DNA methylation involves conversion of unmethylated cytosine to 5-methyl-cytosine (5mC). A proposed method for DNA demethylation involves sequential oxidation and deamination of 5mC, followed by base excision repair1.
A second epigenetic mechanism implicated in learning and memory involves modification of histones, the proteins that DNA wraps around for organization and packaging within the nucleus3,7. The basic organizational unit of the genome is a nucleosome, which is a short segment of DNA wound around a complex of eight histone subunits. Each of these histones has a tail region which can be extensively modified to alter the interactions between histones and DNA and affect the three dimensional structure of chromatin, which in turn can affect transcription. Modifying histones can affect transcription by altering the structure of chromatin and by facilitating interactions with accessory proteins that recognize modified histones. Thus, histone modifications generally represent a more complex and diverse mechanism to affect gene transcription than DNA methylation. In fact, one effect of DNA methylation can be to recruit proteins to mediate histone modification1.
Some major examples of histone modifications are methylation, phosphorylation, acetylation, and ubiquitination, which involve addition of the relevant group to specific amino acids on the histone tails1,8. These modifications play an essential role in regulating the activity of genes by recruiting regulatory proteins and altering the accessibility of DNA. Histone acetylation and phosphorylation generally promote gene activation, the effects of histone methylation are bidirectional and complex, and the effects of ubiquitination remain unclear1. Distinct classes of enzymes are known to mediate each of these modifications. For example, histone acetylation and deacetylation are catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively1.
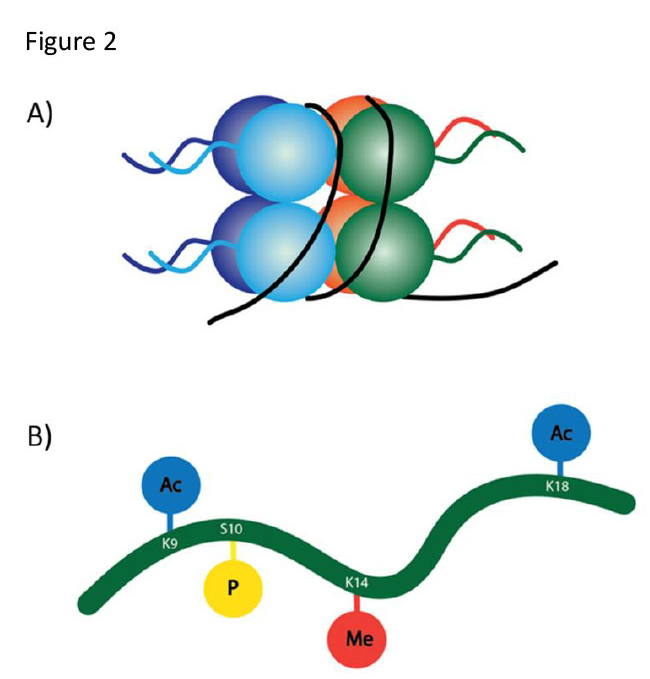
Figure 2 | Nucleosomes and histone tail modifications. A) A nucleosome consists of DNA associated with eight histone proteins, each with a tail domain. B) Along each histone tail, a number of different moieties can be attached to specific amino acids. Acetyl groups (Ac) and methyl groups (Me) are added to lysine (K), and phosphates are added to serine (S)1,7.
The histone modification most studied in learning and memory is histone acetylation3. Compounds that inhibit histone deacetylase enzymes (HDACs) have been shown to enhance certain forms of long term memory formation and synaptic plasticity in vivo9. Despite our knowledge of the role of histone acetylation in learning and memory generally, the specific catalog of genes known to be affected by histone acetylation during learning is relatively sparse. However, some specific cases are well-studied: for example, increased promoter acetylation and expression of brain derived neurotrophic factor (BDNF) in rat prefrontal cortex has been associated with fear conditioning training and extinction10,11.
Histone subunit exchange is a separate type of histone modification beyond addition or removal of functional groups to the tail domains. A full histone complex usually contains two each of histone H2A, H2B, H3, and H4. Beyond these four, there are other isoforms that can be inserted into a nucleosome via a process known as histone subunit exchange1. Only recently has evidence emerged that histone subunit exchange is involved in learning and memory. Last year a group from Dr. David Sweatt’s laboratory at UAB provided the first evidence of dynamic histone subunit exchange as a novel epigenetic mechanism in learning and memory12. They report evidence that the histone variant H2A.z is actively exchanged in the hippocampus and the cortex during fear memory consolidation. Incorporation of H2A.z into a nucleosome is generally associated with absence of DNA methylation and gene activation. Learning-induced recruitment or exclusion of H2A.z from nucleosomes near the transcription start sites of learning and memory genes appears to negatively regulate memory consolidation12.
A role for dynamic epigenetic regulation in at least some forms of learning and memory has been clearly demonstrated. In essence, these mechanisms enable neurons totransiently activate or repress transcription of ensembles of genes that facilitate learning and memory. Epigenetics is a particularly interesting subject for scientists studying learning and memory because it has the potential to underlie both long term and transient changes in gene expression. Additionally, epigenetic dysregulation has been implicated in a wide variety of neurological disorders1,7. Neuroepigenetics promises to be an exciting and productive area of inquiry as we continue to unravel the complex biological underpinnings of learning and memory.
References
- Sweatt, J. D., Meaney, M.J., Nestler, E.J., Akbarian, S. Epigenetic regulation in the nervous system: Basic mechanisms and clinical impact. (Academic Press, 2013).
- Heyward, F. D. & Sweatt, J. D. DNA methylation in memory formation: emerging insights. Neuroscientist, 21(5), 475-489 (2015).
- Peixoto, L. & Abel, T. The role of histone acetylation in memory formation and cognitive impairments. Neuropsychopharmacology, 38(1), 62-76 (2013).
- Miller, C. A. & Sweatt, J. D. Covalent modification of DNA regulates memory formation. Neuron., 53(6), 857-869 (2007).
- Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science, 324(5929),930-935(2009).
- Kriaucionis, S. & Heintz, N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science, 324(5929), 929-930 (2009).
- Tsankova, N., Renthal, W., Kumar, A. &Nestler, E. J. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci., 8(5), 355-367 (2007).
- Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature, 389(6648), 251-260 (1997).
- Levenson, J. M. et al. Regulation of histone acetylation during memory formation in the hippocampus. J Biol Chem., 279(39), 40545-40559 (2004).
- Bredy, T. W. et al. Histone modifications around individual BDNF gene promoters in prefrontal cortex are associated with extinction of conditioned fear. Learn Mem., 14(4), 268-276 (2007).
- Tian, F., Marini, A. M. & Lipsky, R. H. Effects of histone deacetylase inhibitor Trichostatin A on epigenetic changes and transcriptional activation of Bdnf promoter 1 by rat hippocampal neurons. Ann N Y Acad Sci., 1199, 186-193 (2010).
- Zovkic, I. B., Paulukaitis, B. S., Day, J. J., Etikala, D. M. & Sweatt, J. D. Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature, 515(7528), 582-586 (2014).
