Faculty active in this area of research are listed below. For a brief description of their research interests, click on their name in the list. Clicking on the name at the beginning of the brief description links to their detailed personal website.
T. Prescott Atkinson, MD, PhD Research in my laboratory is focused on the role of infection in chronic diseases, especially arthritis and asthma. Ongoing projects in coordination with the UAB Diagnostic Mycoplasma Laboratory are designed to identify mycoplasmas and ureaplasmas in human samples with particular emphasis on the role of those organisms in chronic asthma and extreme prematurity respectively. Previous studies in my laboratory established that Mycoplasma pneumoniae is able to activate mast cells to produce IL-4 through sialic acid-dependent binding to the high affinity receptor for IgE, a finding with potential implications in the pathogenesis of asthma and potentially a general mechanism in the activation of cells of the immune system by that organism. Work is currently proceeding to determine the current prevalence of macrolide resistance strains of M. pneumoniae in the Birmingham area. I am also actively engaged in the development of rational strategies to determine the molecular basis for unidentified immunodeficiencies in patients in my weekly clinical immunology clinics at Children’s of Alabama. Such patients may represent natural “knockouts” or dominant negative mutations in signaling molecules and provide valuable insights into critical steps in receptor signaling in the human immune system.
Peter D. Burrows, PhD Dr. Burrows' laboratory is interested in the development and function of B lymphocytes. Immunoglobulin gene rearrangements, as well as a number of poorly understood changes in gene expression, take place as cells progress through this differentiation pathway. We have been using both cellular and molecular approaches to characterize precursors of human B lineage cells and to identify novel genes whose expression is developmentally regulated. Defects in the expression of such genes could lead to immunodeficiency, whereas inappropriate expression might predispose a cell to malignant transformation. His lab has also begun to explore the function of the multifaceted cytokine, transforming growth factor-beta, in regulating B cell development and function and have identified a novel Fc receptor gene that appears to be expressed in the cytoplasm of germinal center B lymphocytes.
Randy Q. Cron, MD, PhD Host transcription factors exploited by HIV-1. HIV-1, the cause of AIDS, has infected over 40 million individuals world-wide. Although vast improvements in therapy have been developed over the last decade, HIV-1 cannot be totally eliminated from the host due to its ability to enter a resting or latent state in 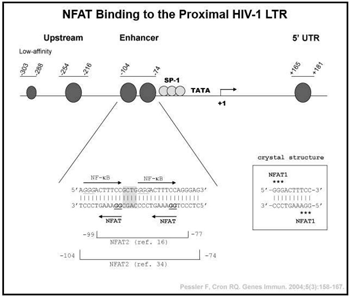 CD4 T cells. Because HIV-1 relies on host transcription factors to replicate, we are exploring the role of the calcium activated nuclear factor of activated T cells (NFAT) transcription factors in regulating HIV-1 transcription. We and others have shown that the CsA-sensitive NFAT proteins bind to the proximal HIV-1 promoter/long terminal repeat (LTR) in vitro and up-regulate HIV-1 transcription. We have further demonstrated that NFAT proteins bind to the integrated HIV-1 LTR in primary human CD4 T cells in vivo by chromatin immunoprecipitation, and this binding is disrupted by the regulatory T cell transcription factor, FOXP3. In addition, we are attempting to exploit NFAT activation as a means of activating HIV-1 LTR activity in latently infected cells. Recently, we identified a novel binding site for the c-maf transcription factor located adjacent to the proximal NFAT sites in the HIV-1 LTR. Our studies reveal synergistic transcriptional activation and increased infection of HIV-1 by c-maf, NFAT2, and NFΚB p65 in primary human IL-4-producing CD4 T cells. Thus, c-maf will likely be a novel therapeutic target in the treatment of HIV-1.
CD4 T cells. Because HIV-1 relies on host transcription factors to replicate, we are exploring the role of the calcium activated nuclear factor of activated T cells (NFAT) transcription factors in regulating HIV-1 transcription. We and others have shown that the CsA-sensitive NFAT proteins bind to the proximal HIV-1 promoter/long terminal repeat (LTR) in vitro and up-regulate HIV-1 transcription. We have further demonstrated that NFAT proteins bind to the integrated HIV-1 LTR in primary human CD4 T cells in vivo by chromatin immunoprecipitation, and this binding is disrupted by the regulatory T cell transcription factor, FOXP3. In addition, we are attempting to exploit NFAT activation as a means of activating HIV-1 LTR activity in latently infected cells. Recently, we identified a novel binding site for the c-maf transcription factor located adjacent to the proximal NFAT sites in the HIV-1 LTR. Our studies reveal synergistic transcriptional activation and increased infection of HIV-1 by c-maf, NFAT2, and NFΚB p65 in primary human IL-4-producing CD4 T cells. Thus, c-maf will likely be a novel therapeutic target in the treatment of HIV-1.
Genetic defects in lymphocyte cytolysis in macrophage activation syndrome. Macrophage activation syndrome (MAS) is a hyper-inflammatory immune response in children and adults that is often triggered by certain infectious (e.g. EBV), autoimmune (e.g. lupus), autoinflammatory (e.g. Still disease), and oncologic (e.g. T cell leukemia) disorders. MAS results in pro-inflammatory cytokine storm leading to pancytopenia, coagulopathy, central nervous system dysfunction, and multi-organ system failure. MAS is frequently lethal like its cousin disease familial hemophagocytic lymphohistiocytosis (fHLH). fHLH is uniformly fatal if not treated aggressively and typically presents in the first few months of life in infants 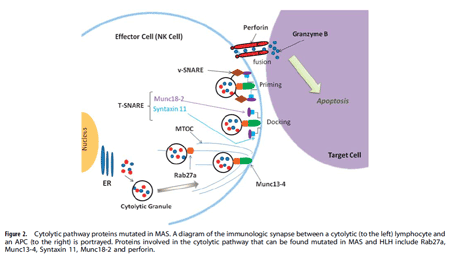 with bi-allelic genetic defects in one of the proteins involved in perforin mediated cytolysis by natural killer (NK) cells and CD8 cytotoxic lymphocytes. Recently, mono-allelic (heterozygous) mutations in cytolytic pathway proteins (e.g. perforin, Munc13-4, Rab27a, etc.) have been identified in a substantial percentage of MAS patients presenting beyond infancy. In our MAS patient cohort, we have identified several mutations, including novel mutants, in a variety of cytolytic pathway genes. Using lentiviral transduction of mutant and wild-type genes into NK cells, we demonstrate decreased cytolytic activity by over-expression of the mutant genes, suggesting a partial dominant-negative effect. These studies suggest that there are likely genetic predispositions to develop MAS, and we are currently exploring the novel mutations and their pathophysiological consequences on lymphocyte mediated cytolytic function.
with bi-allelic genetic defects in one of the proteins involved in perforin mediated cytolysis by natural killer (NK) cells and CD8 cytotoxic lymphocytes. Recently, mono-allelic (heterozygous) mutations in cytolytic pathway proteins (e.g. perforin, Munc13-4, Rab27a, etc.) have been identified in a substantial percentage of MAS patients presenting beyond infancy. In our MAS patient cohort, we have identified several mutations, including novel mutants, in a variety of cytolytic pathway genes. Using lentiviral transduction of mutant and wild-type genes into NK cells, we demonstrate decreased cytolytic activity by over-expression of the mutant genes, suggesting a partial dominant-negative effect. These studies suggest that there are likely genetic predispositions to develop MAS, and we are currently exploring the novel mutations and their pathophysiological consequences on lymphocyte mediated cytolytic function.
Zdenek Hel, PhD Innate immune regulatory activity and neutrophil dysregulation as a driving mechanism of pathogenesis in HIV-1-infection. . Recent evidence demonstrates that neutrophils, the most abundant nucleated immune cell population in the body, play an important role in the regulation of adaptive and innate immune systems. We have shown that neutrophils from HIV-1-infected individuals display an activated phenotype, specific transcriptional profile, and increased rate of degranulation. We propose that HIV-1 infection is associated with altered myeloid cell homeostasis resulting in changes in the population frequency and functional activity of diverse granulocytic populations. Dysregulation of granulocytic recruitment, function, and clearance contributes to the pathogenesis of cardiovascular and liver diseases associated with HIV-1 infection. Specifically, neutrophils in the blood of HIV-1-infected individuals express high levels of PD-L1 that is induced by HIV-1 virions and products of microbial translocation including lipopolysaccharide (LPS). Neutrophil PD-L1 levels correlate with the expression of PD-1 on CD4+ and CD8+ T cells, elevated levels of neutrophil degranulation markers in plasma, and increased frequency of low density neutrophils expressing the phenotype of granulocytic myeloid-derived suppressor cells (G-MDSCs). Neutrophils purified from the blood of HIV-1-infected patients suppress T cell function via several mechanisms including PD-L1/PD-1 interaction and production of reactive oxygen species (ROS). The accumulated data suggest that chronic HIV-1 infection results in an induction of immunosuppressive activity of neutrophils characterized by high expression of PD-L1 and an inhibitory effect on T cell function. This newly identified mechanism of immune suppression mediated by neutrophils may alter our understanding of HIV-1 pathogenesis. Furthermore, we have shown that neutrophils from HIV-1-infected individuals display high capacity to undergo NETosis. Production of neutrophil extracellular traps (NETs) likely contributes to increased risk of cardiovascular and liver diseases in HIV-1-infected individuals.
Neutrophils and cancer. Our research focuses on neutrophils and granulocytic myeloid-derived suppressor cells (G-MDSCs), cell populations that have been recently identified to play a critical role in the regulation of adaptive and innate immune responses in cancer and chronic inflammatory conditions. Production of neutrophil extracellular traps (NETs) by neutrophils contributes to increased risk of cardiovascular and liver disease in cancer patients.
The impact of hormonal contraceptives on HIV-1 acquisition and transmission. Safe and effective methods of contraception represent a critical component of preventive health care reducing maternal and infant mortality, especially in women living in resource-limited settings. Depot medroxyprogesterone acetate (DMPA; Depo-Provera) is a highly effective progestin-based contraceptive and one of the most commonly used contraceptives in sub-Saharan Africa. Several epidemiological studies indicate an association between the use of DMPA and an increased risk of HIV-1 infection. Modelling studies indicate that the use of injectable contraceptives may be responsible for hundreds of thousands of new HIV-1 transmissions annually. It is therefore critically important to identify safe forms of contraception without a significant deleterious effect on systemic and mucosal immune environment. We demonstrated that medroxyprogesterone acetate (MPA) suppresses antigen- immune function of T cells and dendritic cells via direct and indirect mechanisms and increases the rate of HIV-1 proliferation. In a clinical study performed at UAB, we analyzed vaginal biopsies and various immune parameters in the blood of women using various forms of hormonal contraceptives. We showed that the use of MPA is associated with thinning of vaginal epithelial wall and decreased production of IFN-α by plasmacytoid dendritic cells. We have shown that MPA reduces defense mechanisms of genital epithelium by suppression of factors critical for the barrier function and structural integrity of the vaginal and cervical epithelium. Decreased production of these factors reduces the resistance of genital epithelial tissue to microabrasions and increases the probability of HIV-1 transcytosis and transmigration leading to an exposure of target cells in the parabasal epithelium and lamina propria. Furthermore, DMPA and NuvaRing (etonogestrel) significantly suppress the cervicovaginal levels of principal anti-HIV-1 inhibitory factors human β-defensin 2 and 3 and secretory leukocyte protease inhibitor (SLPI). In a recent randomized clinical study in Lusaka, Zambia, we showed that administration of MPA decreases the production of several factors in the cervicovaginal fluid of HIV-1-infected women that may contribute to higher shedding of the virus and potentially to increased rates of viral transmission. In search for safe contraceptives, we have demonstrated that norethisterone (NET) and levonorgestrel (LNG) do not inhibit the function of dendritic cells and T cells and therefore represent safe potential alternative to DMPA.
Harry Schroeder, MD, PhD Ultimately, it is the identity and specificity of the lymphocyte antigen receptor that determines the nature of the immune response to antigen. The mechanisms that underlie the diversification of the B- and T-cell antigen receptor repertoires appear to generate receptor diversity at random. However, repeated examples of near to absolute identity of receptor sequences between individuals suggest the existence of genetically programmed constraints that may be designed to bias the immune system to produce preferred, and perhaps optimal, repertoires. The implication is that violation of these programs could lead to immune dysfunction, and thus to disease. To test this hypothesis, we are developing mouse models wherein we force expression of altered, polyclonal repertoires that violate normal constraints on antigen receptor sequence or structure. In the first of these mice, where we have forced expression of arginine, histidine and asparagine in the HCDR3 interval of immunoglobulin H chains, we observed somatic selection against antigen binding sites that contained an excess number of these charged amino acids, yet the system ultimately failed to recapture the tyrosine and glycine residues normally encoded by wild-type germline sequence. B-cell development was impeded, immunity to influenza virus was impaired, and expression of IgG anti-DNA antibodies was enhanced. These results support the view that optimal distinction between self and non-self is a product of evolutionary selection.