 Plant diseases cause billions of dollars in annual economic losses in the United States. These diseases reduce yields, decrease nutritional value and sometimes contaminate food and feed with toxic compounds. Yet growers are only partially able to control the pathogens that attack their crops, even though they spend millions of dollars combating them.
Plant diseases cause billions of dollars in annual economic losses in the United States. These diseases reduce yields, decrease nutritional value and sometimes contaminate food and feed with toxic compounds. Yet growers are only partially able to control the pathogens that attack their crops, even though they spend millions of dollars combating them.
Now a new discovery from a University of North Carolina at Chapel Hill research team that included current University of Alabama at Birmingham research assistant professor M. Shahid Mukhtar, Ph.D., may finally give plants an advantage over their attackers.
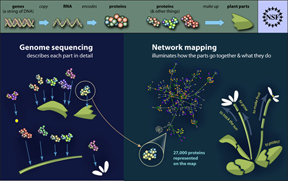
The findings are published in the July 29, 2011, issue of the journal Science. The study suggests that while pathogens employ a diverse arsenal of weapons, they attack only a limited number of cellular targets. Researchers discovered this by building the first comprehensive plant immune “interactome” – a map of the tens of thousands of interactions that link proteins from pathogens and host.
“This is the first-ever immune map of any organism,” says Mukhtar, research assistant professor with the UAB Department of Biology and the lead author of this study. “This discovery will help us understand the mechanism of how pathogens attack their host and how plants fight back and trigger immune responses. We are confident the knowledge we gained from this study will help us prevent plant diseases.”
The study mapped one-third of the interactome of proteins encoded by the genome of the plant Arabidopsis thaliana, or thale cress, a model plant. Arabidopsis has traits that make it useful for understanding the immune mechanisms in other plant species.
“In our lab at UAB, we are characterizing a few key host target proteins using genetics and genomics tools,” says Mukhtar. “We observed both enhanced disease resistance and enhanced disease susceptibility in the mutants of these genes, suggesting the importance of these proteins in resistance to various pathogens.”
The success Mukhtar had in building the plant immune interactome gave him clues of how pathogens can hijack cellular machinery to cause disease — and confidence to build the same sort of model for humans. It is expected that human pathogens also may attack on limited but critical cellular proteins. A human immune interactome will reveal how human immune receptors can activate immune responses.
For this future study Mukhtar will work with Robert Kimberly, M.D., who is senior associate dean for research in the School of Medicine and Howard L. Holley professor and director of the Arthritis, Musculoskeletal and Autoimmunity Center at UAB. Kimberly also is program director for the Multi-disciplinary Clinical Research Center (MCRC) funded by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), as well as a professor of microbiology and senior scientist for the UAB Comprehensive Cancer Center.
“The success of this study shows how valuable model organisms can be in understanding fundamental principles in biology,” says Kimberly. “We are now looking to build a human interactome at UAB in our continuing efforts to fight diseases such as arthritis, diabetes, Crohn’s and cancer.”
The interdisciplinary research between Mukhtar and Kimberly into a human interactome will begin as soon as they obtain funding. Their work will be performed in collaboration with Pascal Braun, Ph.D., David Hill, Ph.D., and Marc Vidal, Ph.D., all with the Dana Farber Cancer Institute in Boston, Mass.
“Just to get where we are today we did robotic screening of millions of combinations – at least over 12 million,” says Mukhtar. “In all that work over the course of five years I never got frustrated or lost hope because you could see an interactome map developing that may provide a path to a life-changing discovery.”
The original study, “Independently Evolved Virulence Effectors Converge onto Hubs in a Plant Immune System Network,” was co-written by researchers from over a dozen institutions. Along with Mukhtar, the lead authors were Jeff Dangl, Ph.D., John N. Couch professor of biology in the UNC College of Arts and Sciences, HHMI-GBMF Investigator, a member of the Carolina Center for Genome Sciences and an adjunct professor of microbiology and immunology in the UNC School of Medicine; Petra Epple, research associate in Dangl’s lab; and Anne-Ruxandra Carvunis and Matija Dreze from the Dana-Farber Cancer Institute in Boston. Additional authors from the Dangl lab were research associate Marc Nishimura, graduate student Yijian He and undergraduates Nathan McDonald and Christopher Harbort.