Bioinformatics
Data Analysis Services Offered:
- Next Generation Sequence (NGS) Analysis
- Causal Variant Analysis
- System Biology Analysis
Now that you have your raw data (whether it is some type of Next Generation Sequencing, legacy chip-based arrays, or from other omics platforms), the big question arises: “What type of analyses needs to be performed to make sense of it?” We offer a variety of tools necessary to answer your questions.
Please contact Dr. David Crossman for all of your data analysis needs. Dr. Crossman provides expertise in both the data analysis and the downstream applications of Systems Biology. He is proficient with a variety of data analysis tools via the command line and the R statistical package software. He is also proficient in a variety of Systems Biology tools such as Ingenuity Pathway Analysis (IPA), ThomsonReuters' Metacore, and Cytoscape. He performs systems analyses on various –omics datasets obtained from the various –omics platforms available to any investigator.
Not only will Dr. Crossman analyze your Big Data, but he will help you with study design (before you even start your project), review your grants that require Bioinformatics expertise, provide biological insight into your hypothesis for manuscript/grant preparation, produce figures/tables for your manuscripts/grants, and review your manuscripts.
Next Generation Sequencing Analysis
There is an abundance of tools available for Next Generation Sequencing analysis via command line. Dr. Crossman uses the High Performance Computing (HPC) cluster here at UAB, called Cheaha, to analyze all of the Big Data that arises from NGS and other –omics platforms. Here are only a few examples of NGS analysis and some analysis tools performed here at the Core (if we listed them all, this page would go on and on and on, that you would stop reading this page):
| Type of –Seq** |
Description |
Analysis Tools** |
| RNA-Seq |
Identify expression levels of genes/isoforms, gene fusion, mutations/SNPs, alternative gene spliced transcripts, and more. |
STAR, kallisto, salmon, RSEM, HISAT2, HTSeq-count, subreads featureCount, DESeq2, edgeR, Stringtie/Ballgown |
| Whole genome/exome |
Identify SNPs, INDELs and structural variants |
BWA, Bowtie2, GATK, Freebayes, Lumpy |
| ChIP-Seq |
Identify the binding sites of DNA-associated proteins |
Bowtie2, MACS2, Homer |
| ATAC-Seq |
Identify chromatin accessibility |
Bowtie2, MACS2, Homer |
| Methylation |
Identify hyper/hypo-methylated regions of the DNA (CpG Islands, shores, shelves, etc…) |
Bismark, methylKit, bsseq, bis-snp |
| microRNA-Seq |
Identify microRNAs that modulate protein epxression through transcript degradation, inhibition of translation, or sequestering transcripts |
CAP-miRSeq, miARma-Seq |
| Single-cell RNA-Seq# |
Examine the sequence information from individual cells providing a higher resolution of cellular differences and a better understanding of the function of an individual cell in the context of its microenvironment |
STAR, kallisto, salmon, scater, Seurat |
**If you don’t see your favorite –Seq or Analysis Tool listed, please contact Dr. Crossman as he has expertise in a lot more than these brief examples listed. # Currently working on an analytical pipeline, so testing a wealth of tools.
Causal Variant Analysis
A question asked by those performing whole genome/exome sequencing is “What disease-related causal variant(s) are associated with the phenotype I’m studying?” Identifying the causal variant(s) can lead to further studies of gene targets to better characterize disease mechanisms. For the Undiagnosed Disease Program (UDP), we use Agilent’s Cartagenia Bench Lab NGS to identify and report the structural and molecular variants (including tumor/normal) associated with a patient’s phenotype. We also have access to Ingenuity Variant Analysis, which aids in the identification of disease-causing variants.
Systems Biology Analysis
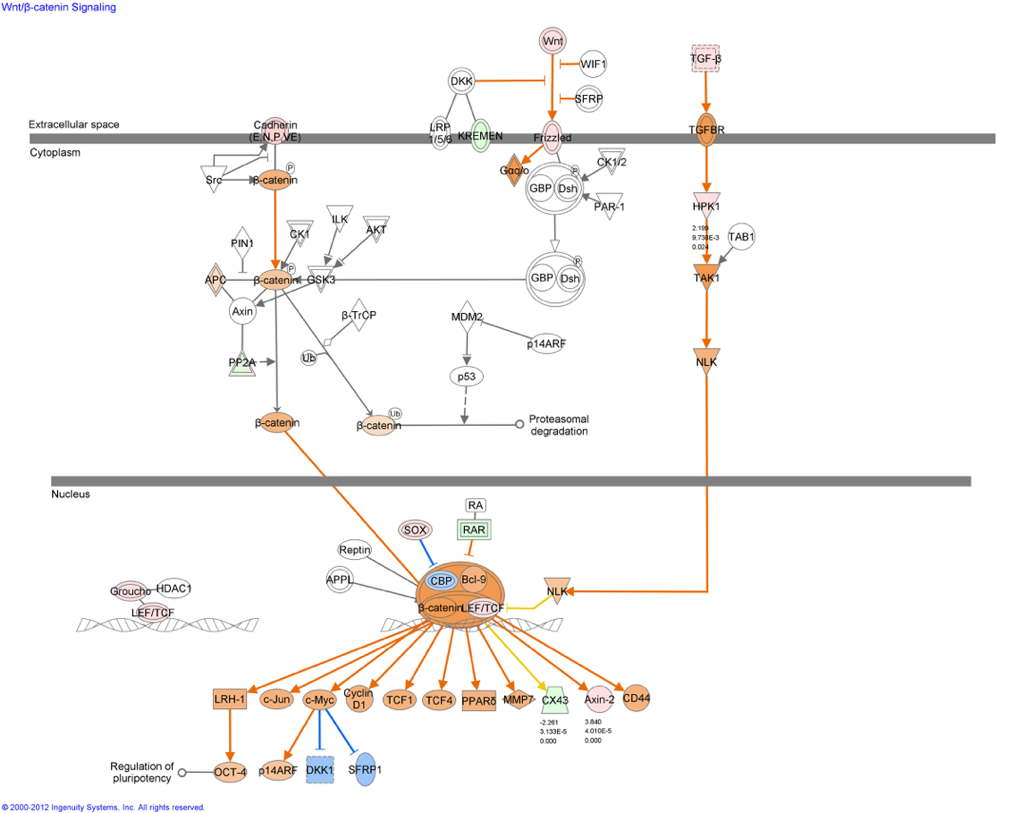
The next question that arises from all of this Big Data is, “What do I do now with my list of differentially expressed genes/microRNAs?” Performing Systems Biology on these lists will allow you to gain insight into the biological meaning of these lists. One of the several Systems Biology tools we use is Ingenuity Pathway Analysis.
Ingenuity IPA: "IPA is software that helps researchers model, analyze, and understand the complex biological and chemical systems at the core of life science research. IPA has been broadly adopted by the life science research community and is cited in thousands of peer-reviewed journal articles."
"IPA helps you understand biology at multiple levels by integrating data from a variety of experimental platforms and providing insight into the molecular and chemical interactions, cellular phenotypes, and disease processes of your system. Even if you don’t have experimental data, you can use IPA to intelligently search the Ingenuity® Knowledge Base for information on genes, proteins, chemicals, drugs, and molecular relationships to build biological models or get up to speed in a relevant area of research. IPA provides the right biological context to facilitate informed decision-making, advance research project design, and generate new testable hypotheses."