Media contact: Bob Shepard
 Human biology is full of surprises — especially for drug makers. Viagra wasn’t designed for erectile dysfunction. Rogaine didn’t start out as a hair-loss cream. Both drugs were meant to treat cardiovascular issues (as sildenafil and minoxidil, respectively), until patients reported their sexual and follicular side effects.
Human biology is full of surprises — especially for drug makers. Viagra wasn’t designed for erectile dysfunction. Rogaine didn’t start out as a hair-loss cream. Both drugs were meant to treat cardiovascular issues (as sildenafil and minoxidil, respectively), until patients reported their sexual and follicular side effects.
When his son was diagnosed with an ultra-rare disease, computer scientist Matt Might, Ph.D., kicked off a search for answers. His quest led to partnerships with researchers across the country, a White House appointment, a faculty position at Harvard, and a profile in the New Yorker. It also led to the discovery that off-the-shelf drugs such as Prevacid could bring remarkable off-label benefits to patients like Might’s son, Bertrand. Prevacid, it turned out, appears to have helped improve Bertrand’s developmental trajectory and eliminated his chronic lung infections.
Related stories:The Power of One — read more about precision medicine at UAB in these stories from UAB Magazine. |
Now, as the director of the UAB Precision Medicine Institute, Might and a team of collaborators from the PMI, the UAB Informatics Institute, and California’s Scripps Research Institute are building a software engine designed to generate similar breakthrough discoveries for patients around the world. It’s called mediKanren. (“Kanren” is based on a Japanese word meaning “connection.”) In addition to sparking new research ideas, it could connect patients with potential treatments that are already FDA-approved for another condition.
| "The NIH has a big problem," explains Will Byrd. "There are hundreds of databases out there filled with genes, drugs, phenotypes, proteins, disease symptoms, whatever you can imagine." But how to unlock access to all those findings? That's where tools like mediKanren come in. |
Finding new connections
“It’s a reasoning engine for biomedical knowledge,” Might says. Or, more colorfully, an artificial intelligence designed to hunt out new treatments with the logical powers of Spock, the deductive acumen of Sherlock Holmes, and the medical genius of TV’s Dr. House. It understands the arguments made in scientific research — X inhibits Y, Y causes Z — and can draw logical conclusions between them. (See “How It Works.”) The scratch-built program debuted at an NIH-sponsored “hackathon” in January. With a $600,000 grant, the team is now taking mediKanren to the next level.
“The NIH has a big problem,” explains computer scientist Will Byrd, Ph.D., a longtime Might collaborator now at UAB, and one of the principal developers of miniKanren, the logical reasoning tool that inspired mediKanren. (The proof-of-concept code for mediKanren is available for download on GitHub.) “There are hundreds of databases out there filled with genes, drugs, phenotypes, proteins, disease symptoms, whatever you can imagine.” The NIH’s National Center for Advancing Translational Sciences (NCATS) launched the Biomedical Data Translator program to develop standard formats that would let researchers ask questions across a wide range of data sources. But unlocking access to these findings is only one step. Is it possible to design tools that could use the data to answer important translational research questions?
| Crucially, mediKanren doesn’t just return results, but also provides “an explanation for why they might work,” says Might. |
"Iron Chef" for science
That question sparked the NCATS Reasoning Engine project and the San Diego hackathon. The competition brought together teams from across the country in an “Iron Chef”-style competition, in which the organizers revealed a disease after the teams arrived. Each team’s software had to look through the medical literature and find new treatments on the spot.
“In an hour and a half, our tool generated the top two candidates in clinical trials right now, plus 10 more potential treatments,” says Might. Based on those findings, researchers studying Fanconi’s anemia have started a dialogue with drug companies. Crucially, mediKanren doesn’t just return results, but also provides “an explanation for why they might work,” says Might. Users can follow mediKanren’s thought processes, seeing the chain of research papers and other clues the program followed to arrive at those conclusions. “Logical reasoning is good at giving explanations,” Byrd says.
It’s possible that a digital investigator like mediKanren could run continually, he adds – churning through new research to find fresh connections, and alerting its human counterparts whenever it comes across something promising.

Testing, testing
At the moment, Might is stress-testing mediKanren in the real world. As he consults with patients and researchers at UAB, Might enters search terms on his laptop, generating a host of new hypotheses in a single meeting. Gene sequencing might reveal that a patient’s genetic mutation is causing overproduction of a specific protein, for example. Might can use mediKanren to find any FDA-approved drugs that inhibit that protein.
“It’s fast enough so that you can do many queries in an hour-long consultation,” Byrd says. And even if the search turns up nothing today, it could be automated to run every week, he adds — constantly querying the medical literature for new insights. After these sessions, Might shares feedback with Byrd and the other members of the mediKanren team, including Greg Rosenblatt of the Precision Medicine Institute; James Cimino, M.D., and Jake Chen, Ph.D., director and co-director of UAB’s Informatics Institute; and Andrew Su, Ph.D., and colleagues at the Scripps Research Institute in La Jolla, California.
With their NCATS funding, the team is refining their original proof-of-concept design. Eventually, it could combine the best of the machine learning approach with mediKanren’s logic-based insights.
“The goal is to reduce the cost of drug discovery and repurposing through modern software engineering,” Byrd says. Adds Might: “There isn’t anything else like it.”
How It Works
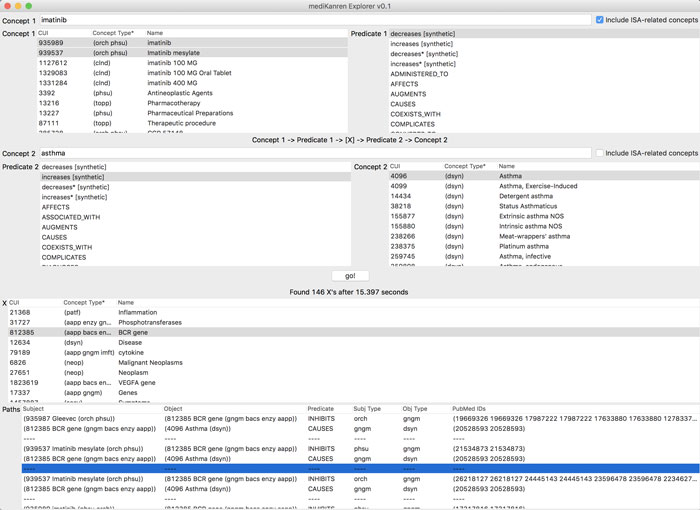
mediKanren (sample screenshot above) is programmed to find hidden connections in biomedical data by seeking out logical relationships between concepts.
 Type in two concepts and a predicate — for example, a disease and a possible new treatment option.
Type in two concepts and a predicate — for example, a disease and a possible new treatment option.
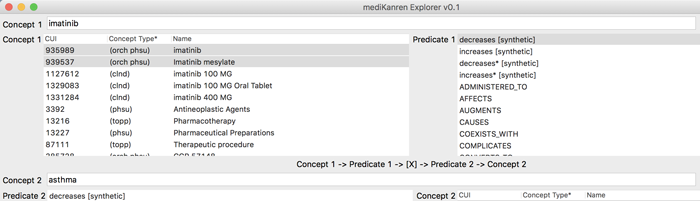
 mediKanren will search through 97 million assertions, taken from more than 20 million scientific papers. (Updates are adding more databases, including a full list of FDA-approved medications.)
mediKanren will search through 97 million assertions, taken from more than 20 million scientific papers. (Updates are adding more databases, including a full list of FDA-approved medications.)
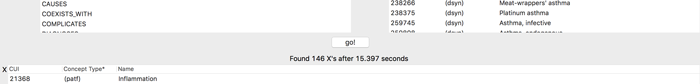
 In 15 seconds, the program highlights several possible connections, serving up links to the relevant papers on PubMed, the NIH’s vast research database.
In 15 seconds, the program highlights several possible connections, serving up links to the relevant papers on PubMed, the NIH’s vast research database.
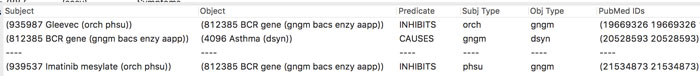
 mediKanren may find tens of thousands of possible links to explore, but it is carefully designed to highlight the top results for users. “If people don’t find something they’re interested in within the first five results, they’ll give up,” says Byrd.
mediKanren may find tens of thousands of possible links to explore, but it is carefully designed to highlight the top results for users. “If people don’t find something they’re interested in within the first five results, they’ll give up,” says Byrd.
This research is supported by the National Center For Advancing Translational Sciences of the National Institutes of Health under Award Number OT2TR002517. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.