Arguably the most prominent target for drug development these days is kinase-directed therapy. Almost daily, there are news reports, articles, patents, and clinical trials describing how kinase inhibitors are changing how medical therapy is carried out and improving patient outcomes.
The reality, however, is that proper patient and drug selection has remained elusive. Indeed, it is apparent that kinase-targeted agents show good efficacy in a subset of patients, yet it is not entirely clear why one group will respond while others fail. In some instances, there are known genetic mutations that may predict for response – such as when a kinase is overexpressed in an activated form (e.g., EGFR mutations in lung cancer or B-Raf mutations in melanoma) but often these mutations are rare events. In many cases, kinase targeted agents are used in completely unselected patients without any biological testing to inform the clinician.
Part of the reason for this lack of patient selection is due to the fact that the molecular tests that are utilized to search for a biomarker of response are often far removed from the target of the therapeutic. Specifically, kinase inhibitors, by definition, target activated protein enzymes (kinases), yet almost all biomarker studies investigate DNA or RNA, which may have no correlation to protein levels. Moreover, protein levels do not necessarily correlate to activity either. In other words, differences in kinase activity, are often “invisible” to genomic (DNA), transcriptomic (RNA), and most proteomic (protein) platforms because kinases are predominantly regulated post-translationally (i.e., after being made into proteins). As such, platforms that look at the function of proteins, so-called “functional proteomics,” are desperately needed for kinase-targeted therapies. The Kinome Core represents a functional proteomic approach with a very unique advantage.
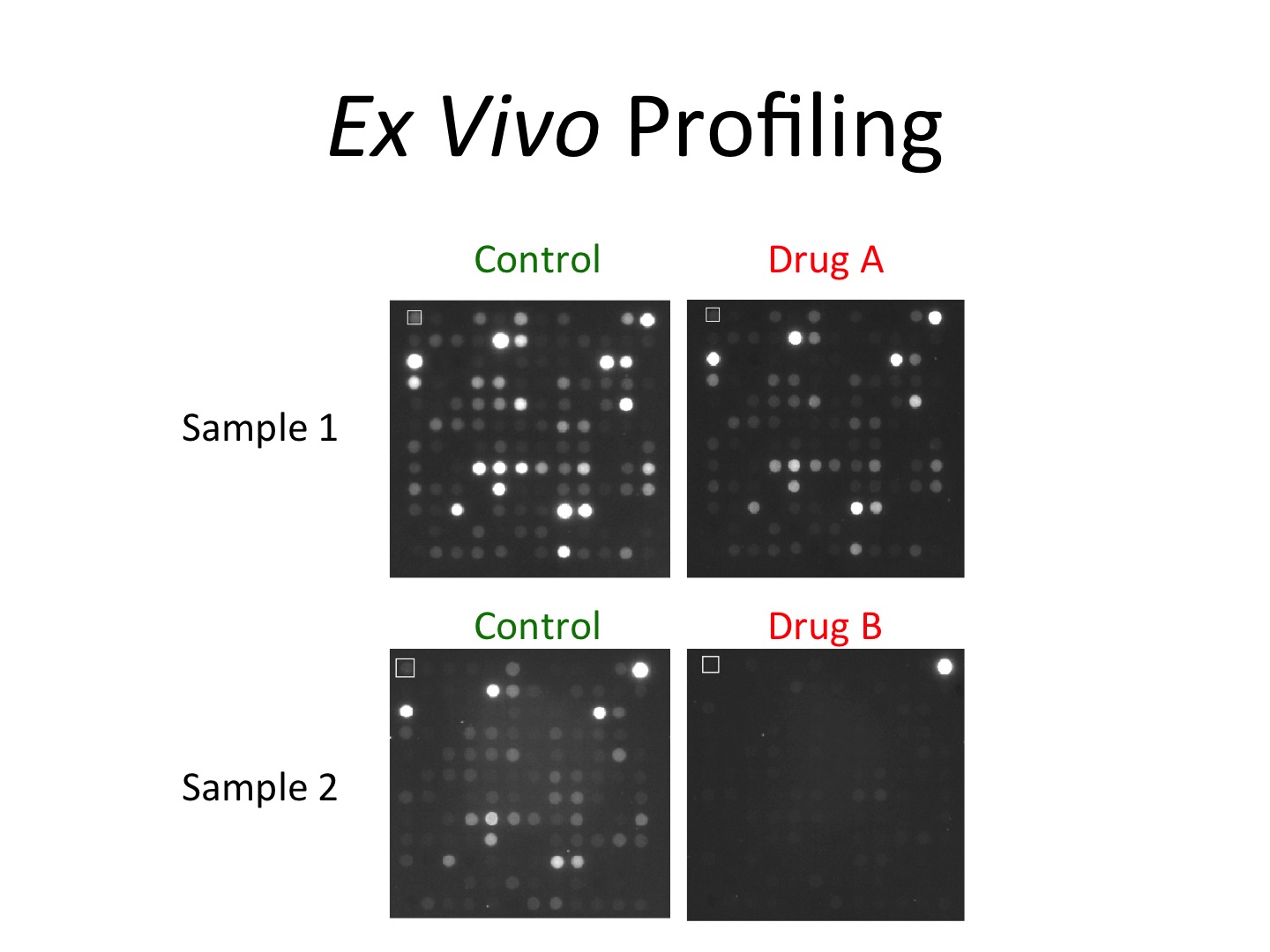
One of the real advantages of the Kinome Core platform is that because the platform directly measures kinase enzymatic activity in samples, it is also possible to block the enzymatic activity using small molecule inhibitors. In other words, the Kinome Core can “challenge” the sample’s kinase activity with a kinase inhibitor to measure the impact for that inhibitor in terms of kinase function, which we call “ex vivo profiling.” This provides an additional level of information in an otherwise “static” sample. An example of this is shown in the adjacent figure. The kinomic “fingerprint” of the tumor specimen changes based on which kinase inhibitor is added to the mix. It should be noted that the ability for the naked eye to distinguish the changes is much less than the imaging software that is used in the Kinome Core, so a great deal of information can be gleaned using our system. More relevant biomarkers can be produced in this manner because the functional test is truly examining the target of the therapeutic. As such, we envision the use of ex vivo profiling will one day help clinicians select proper therapies for their patients to usher in true precision medicine.