Neuroscience trainees navigate challenges beyond the lab
By: Vicki Hixon
Published Date: Oct 14
By Nancy Mann Jackson • Photos by Steve Wood
Even though she possessed a bachelor’s degree in chemistry and a master’s in biotechnology, Lillian Brady felt that she didn’t fit into the booming field of neuroscience. “As an underrepresented student, it can be easy to get into the mind frame that you don’t belong,” says Brady, a graduate of Alcorn State University, a historically black university in southwest Mississippi.
 Each Roadmap Scholar works closely with a research mentor. Here, scholar Lillian Brady (right) meets with neurobiology associate professor Lynn Dobrunz.
Each Roadmap Scholar works closely with a research mentor. Here, scholar Lillian Brady (right) meets with neurobiology associate professor Lynn Dobrunz.
But then the Jackson, Mississippi, native found a place where she fit in perfectly: UAB’s Neuroscience Roadmap Scholars program, which is designed to help engage and retain underrepresented graduate trainees—including ethnic minorities and students with disabilities—in the neuroscience workforce.
Brady, now a doctoral student in UAB’s Department of Neurobiology, calls the program a “confidence booster” that has provided both support and encouragement. “I’ve been exposed to scientists who look like me and who have had some of the same challenges I am facing now,” she says. “These scientists are thriving in their fields, so I have no doubt that I can be successful in whatever career path I choose.”
The problem isn’t talent or academic strength; rather, many students from diverse backgrounds or with disabilities may struggle with financial challenges, family needs, or a lack of confidence. Lubin and Lori McMahon, Ph.D., UAB Graduate School dean and UAB Comprehensive Neuroscience Center director, have seen such roadblocks dismantle the plans and goals of numerous students and coworkers.
 Roadmap co-directors Lori McMahon (left) and Farah Lubin (right) serve as career coaches for scholars outside the lab.
Roadmap co-directors Lori McMahon (left) and Farah Lubin (right) serve as career coaches for scholars outside the lab.
To plug the pipeline’s holes, Lubin and McMahon developed Roadmap Scholars to emphasize mentoring, support networks, team-building experiences, and a community of peers to help guide more students to achieve neuroscience careers. Funded by a grant from the National Institute of Neurological Disorders and Stroke, the program launched in 2014. Today it includes 25 doctoral students.
The funding does not pay tuition or stipends; instead, it enhances and provides educational experiences to prepare students for further study and to encourage them to remain in neuroscience. And it works in tandem with the students’ graduate curriculum. For example, every Roadmap Scholar matches with a “career coach” in addition to a primary research mentor. The coach is a faculty member not on the student’s thesis committee who is available to talk about nearly anything—scientific projects, publishing and presenting, conflict management, or life in general, Lubin says. In doing so, these coaches provide additional professional perspectives and serve as an additional layer of support.
For Leland Fleming, a Fort Worth, Texas, native and UAB Graduate Biomedical Sciences doctoral student, the program’s peer and faculty network has made his goal of becoming a neuroscientist more tangible. “With its guidance and inspiration, it has made a once seemingly impossible goal something that I feel more than capable of accomplishing,” he says.
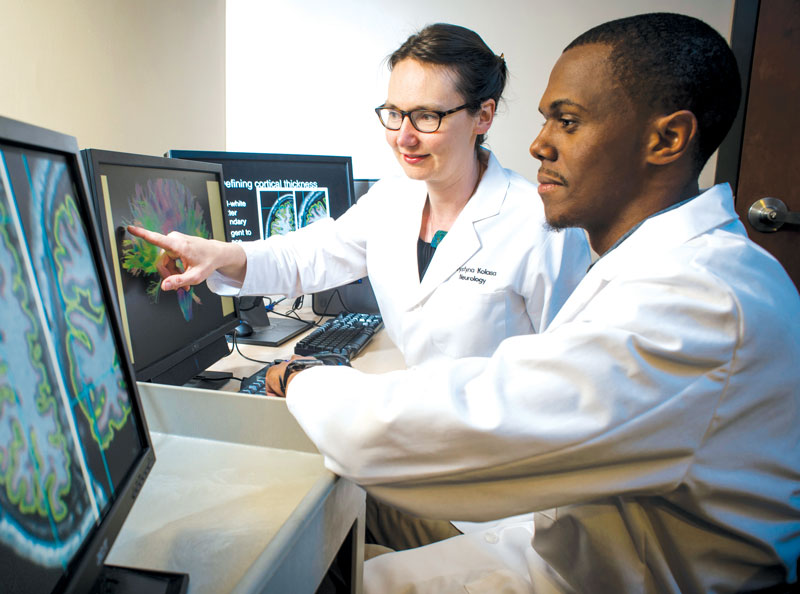 Leland Fleming (right) with his research mentor, neurobiology assistant professor Kristina Visscher
Leland Fleming (right) with his research mentor, neurobiology assistant professor Kristina Visscher
For Fleming, key conference takeaways included insights on common challenges that minority students can face in graduate school. These might be “feelings of inadequacy or feelings that he or she is merely an impostor on the brink of being exposed at any given moment,” he explains. But he and other students also received “terrific advice on coping with these issues when they arise,” Fleming says.
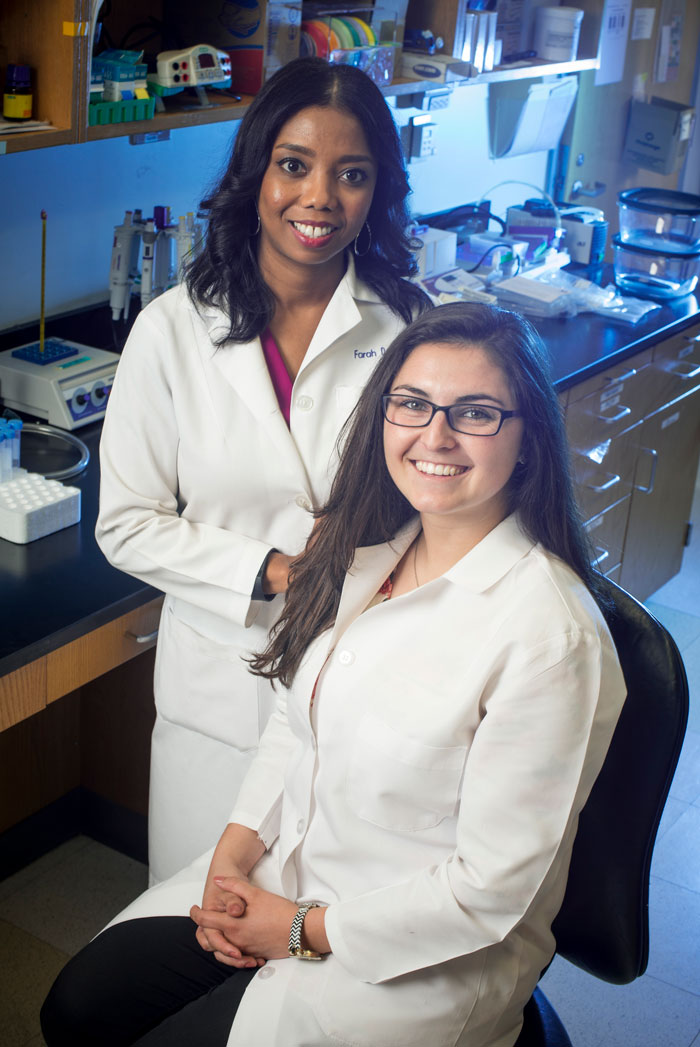 Lubin serves as a research mentor for scholar Rylie Hightower (right).
Lubin serves as a research mentor for scholar Rylie Hightower (right).
The conference also helps to build strong bonds among students. Matthew Timberlake, a second-year graduate student who grew up in Fort Worth and Enterprise, Alabama, has enjoyed helping his colleagues to design and present lectures—even sharing his own research data with them. “I like the sense of community,” he says. “My colleagues and I—especially the upperclassmen—have an important role in discussions,” he says. “We help to ease some of the unknown for the underclassmen. In the same way, I also benefit from the upperclassmen’s advice and guidance.”
Rylie Hightower, an Albuquerque native who came to UAB after earning a nursing degree in New Mexico, is one of the newer students benefiting from those discussions. At a Roadmap spring retreat, Hightower sought advice from older students. Those interactions “will help me throughout my time at UAB and beyond,” she says.
The program has “made me feel at home away from home,” Hightower says. “But it also has helped me understand that a supportive community can greatly contribute to the way I think and perform as a student and as a scientist.”
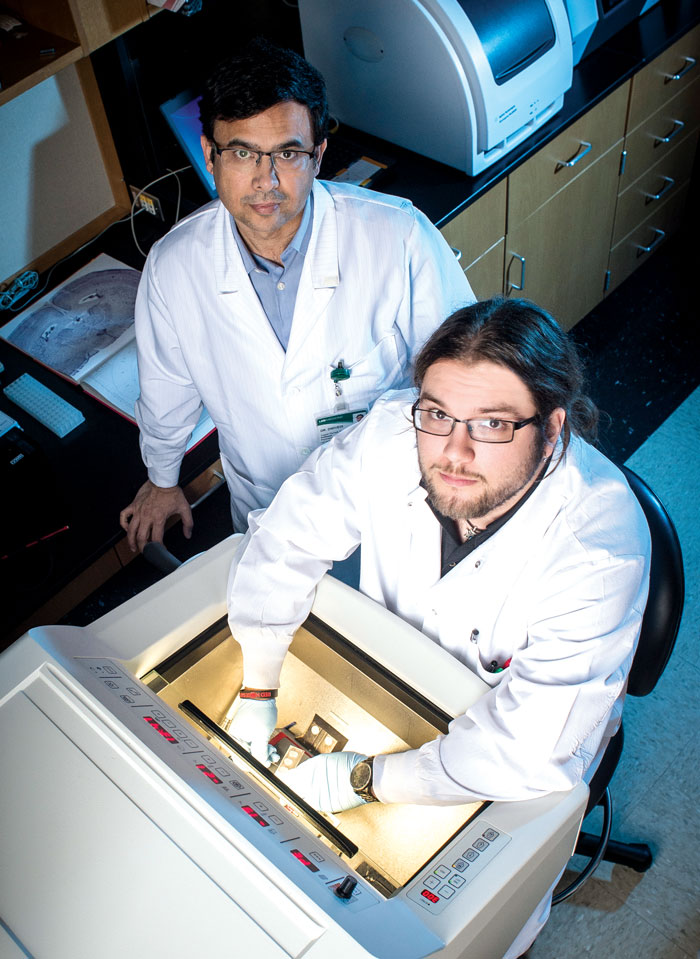 Matthew Timberlake works with research mentor and psychiatry professor Yogesh Dwivedi.
Matthew Timberlake works with research mentor and psychiatry professor Yogesh Dwivedi.
Every student also learns about neuroscience-related job opportunities in business and industry. “Often, students are encouraged to build a career path focused on remaining in academia,” says Megan Rich, a Fairfield, Connecticut, native and UAB Graduate Biomedical Sciences student. “The program has been great about expressing other options after graduate school and that it is OK to think about them.”
By helping these students to feel empowered, capable, and supported, the Neuroscience Roadmap Scholars program will lay the groundwork for future progress in one of the fastest growing scientific fields, McMahon says. Lubin agrees, noting that diversity is crucial for continuing advancements: “Diverse groups can offer unique perspectives and unique ideas, which will push the field forward.”
• Learn more about the Neuroscience Roadmap Scholars Program, including how to apply.
• Give something and change everything for the next generation of neuroscientists by supporting the School of Medicine.
Even though she possessed a bachelor’s degree in chemistry and a master’s in biotechnology, Lillian Brady felt that she didn’t fit into the booming field of neuroscience. “As an underrepresented student, it can be easy to get into the mind frame that you don’t belong,” says Brady, a graduate of Alcorn State University, a historically black university in southwest Mississippi.
 Each Roadmap Scholar works closely with a research mentor. Here, scholar Lillian Brady (right) meets with neurobiology associate professor Lynn Dobrunz.
Each Roadmap Scholar works closely with a research mentor. Here, scholar Lillian Brady (right) meets with neurobiology associate professor Lynn Dobrunz.But then the Jackson, Mississippi, native found a place where she fit in perfectly: UAB’s Neuroscience Roadmap Scholars program, which is designed to help engage and retain underrepresented graduate trainees—including ethnic minorities and students with disabilities—in the neuroscience workforce.
Brady, now a doctoral student in UAB’s Department of Neurobiology, calls the program a “confidence booster” that has provided both support and encouragement. “I’ve been exposed to scientists who look like me and who have had some of the same challenges I am facing now,” she says. “These scientists are thriving in their fields, so I have no doubt that I can be successful in whatever career path I choose.”
Strengthening the Pipeline
“Through our experience working with students, we realized the pipeline for diverse neuroscientists was leaky,” recalls Farah Lubin, Ph.D., associate professor of neurobiology. “We might start with many diverse, motivated students, but somehow, many of them don’t make it into successful careers.”The problem isn’t talent or academic strength; rather, many students from diverse backgrounds or with disabilities may struggle with financial challenges, family needs, or a lack of confidence. Lubin and Lori McMahon, Ph.D., UAB Graduate School dean and UAB Comprehensive Neuroscience Center director, have seen such roadblocks dismantle the plans and goals of numerous students and coworkers.
 Roadmap co-directors Lori McMahon (left) and Farah Lubin (right) serve as career coaches for scholars outside the lab.
Roadmap co-directors Lori McMahon (left) and Farah Lubin (right) serve as career coaches for scholars outside the lab.To plug the pipeline’s holes, Lubin and McMahon developed Roadmap Scholars to emphasize mentoring, support networks, team-building experiences, and a community of peers to help guide more students to achieve neuroscience careers. Funded by a grant from the National Institute of Neurological Disorders and Stroke, the program launched in 2014. Today it includes 25 doctoral students.
The funding does not pay tuition or stipends; instead, it enhances and provides educational experiences to prepare students for further study and to encourage them to remain in neuroscience. And it works in tandem with the students’ graduate curriculum. For example, every Roadmap Scholar matches with a “career coach” in addition to a primary research mentor. The coach is a faculty member not on the student’s thesis committee who is available to talk about nearly anything—scientific projects, publishing and presenting, conflict management, or life in general, Lubin says. In doing so, these coaches provide additional professional perspectives and serve as an additional layer of support.
For Leland Fleming, a Fort Worth, Texas, native and UAB Graduate Biomedical Sciences doctoral student, the program’s peer and faculty network has made his goal of becoming a neuroscientist more tangible. “With its guidance and inspiration, it has made a once seemingly impossible goal something that I feel more than capable of accomplishing,” he says.
 Leland Fleming (right) with his research mentor, neurobiology assistant professor Kristina Visscher
Leland Fleming (right) with his research mentor, neurobiology assistant professor Kristina VisscherCreating a Community
The support system kicks into gear even before the scholars’ first semester at the program’s summer NEURAL (National Enhancement of Underrepresented Academic Leaders) Conference, which draws neuroscience trainees from across the country. There, students can hone their networking and public speaking skills and interact with the field’s academic leaders. For instance, at the 2015 inaugural conference, leading neuroscientist Roger Nicoll, M.D., of the University of California San Francisco drew an emotional response from the audience when he spoke of succeeding despite his lifelong struggle with dyslexia.For Fleming, key conference takeaways included insights on common challenges that minority students can face in graduate school. These might be “feelings of inadequacy or feelings that he or she is merely an impostor on the brink of being exposed at any given moment,” he explains. But he and other students also received “terrific advice on coping with these issues when they arise,” Fleming says.
 Lubin serves as a research mentor for scholar Rylie Hightower (right).
Lubin serves as a research mentor for scholar Rylie Hightower (right).The conference also helps to build strong bonds among students. Matthew Timberlake, a second-year graduate student who grew up in Fort Worth and Enterprise, Alabama, has enjoyed helping his colleagues to design and present lectures—even sharing his own research data with them. “I like the sense of community,” he says. “My colleagues and I—especially the upperclassmen—have an important role in discussions,” he says. “We help to ease some of the unknown for the underclassmen. In the same way, I also benefit from the upperclassmen’s advice and guidance.”
Rylie Hightower, an Albuquerque native who came to UAB after earning a nursing degree in New Mexico, is one of the newer students benefiting from those discussions. At a Roadmap spring retreat, Hightower sought advice from older students. Those interactions “will help me throughout my time at UAB and beyond,” she says.
The program has “made me feel at home away from home,” Hightower says. “But it also has helped me understand that a supportive community can greatly contribute to the way I think and perform as a student and as a scientist.”
 Matthew Timberlake works with research mentor and psychiatry professor Yogesh Dwivedi.
Matthew Timberlake works with research mentor and psychiatry professor Yogesh Dwivedi.The Road Ahead
Career planning is a key component of the program. Brady, who plans to obtain a postdoctoral position following her UAB training, found Roadmap’s “postdoctoral school” to be extremely valuable. She describes it as a weeklong crash course for senior graduate students covering “everything we need to know about postdoctoral positions. Not only were we given pointers on securing the right postdoc for our interest, but we also were exposed to postdocs in fields we might not have considered.”Every student also learns about neuroscience-related job opportunities in business and industry. “Often, students are encouraged to build a career path focused on remaining in academia,” says Megan Rich, a Fairfield, Connecticut, native and UAB Graduate Biomedical Sciences student. “The program has been great about expressing other options after graduate school and that it is OK to think about them.”
By helping these students to feel empowered, capable, and supported, the Neuroscience Roadmap Scholars program will lay the groundwork for future progress in one of the fastest growing scientific fields, McMahon says. Lubin agrees, noting that diversity is crucial for continuing advancements: “Diverse groups can offer unique perspectives and unique ideas, which will push the field forward.”
• Learn more about the Neuroscience Roadmap Scholars Program, including how to apply.
• Give something and change everything for the next generation of neuroscientists by supporting the School of Medicine.
Published October 2016
Visscher Lab Makes News
By: Vicki Hixon
Published Date: Sep 15
UAB researchers use expanded computing power to accelerate big-data science by Jeff Hansen
Computing challenges are found across the UAB campus, from physics and neurology to genetics and the microbiome. Alabama’s most advanced supercomputer is now at UAB, making it possible to solve these challenges.
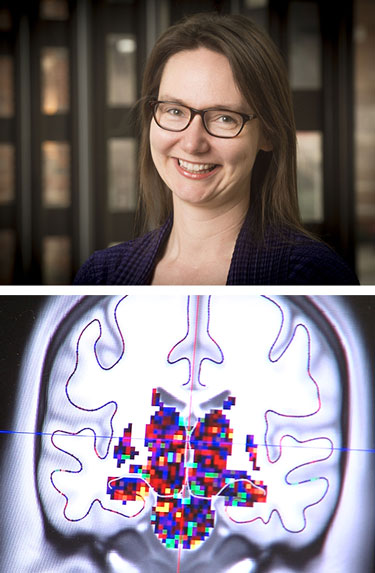 Kristina Visscher is using fMRI images from several hundred brains to learn how the brain adapts after long-term changes in vision.What do the human brain, the 3 billion base-pair human genome and a tiny cube of 216 atoms have in common?
Kristina Visscher is using fMRI images from several hundred brains to learn how the brain adapts after long-term changes in vision.What do the human brain, the 3 billion base-pair human genome and a tiny cube of 216 atoms have in common?
All of them, from the tiny cube to the 3-pound human brain, create incredibly complex computing challenges for University of Alabama at Birmingham researchers, and aggressive investments in UAB’s IT infrastructure have opened new possibilities in innovation, discovery and patient care.
For example, Kristina Visscher, Ph.D., assistant professor of neurobiology, UAB School of Medicine, uses fMRI images from several hundred human brains to learn how the brain adapts after long-term changes in visual input, such as macular degeneration. Frank Skidmore, M.D., an assistant professor of neurology in the School of Medicine, studies hundreds of brain MRI images to see if they can predict Parkinson’s disease.
David Crossman, Ph.D., bioinformatics director in UAB’s Heflin Center for Genomic Science, deciphers the sequences of human genomes for patients seeking a diagnosis in UAB’s Undiagnosed Diseases Program, and he processes DNA sequencing for UAB researchers who need last-minute data for their research grant applications.
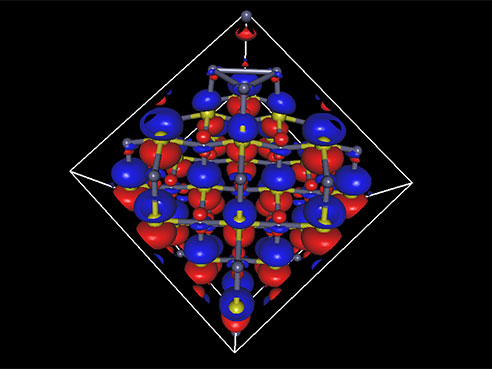
Ryoichi Kawai, Ph.D., an associate professor of physics in the UAB College of Arts and Sciences, is laying the groundwork for a better infrared laser by calculating the electronic structure for a cube made up of just 216 atoms of zinc sulfide doped with chromium or iron.
Each researcher faces a mountain range of computational challenges. Those mountains are now easier to scale with UAB’s new supercomputer — the most advanced in Alabama for speed and memory.
Using 1,000 cores, it would take two to three months of continuous computing to solve the structure, Kawai says. Both the increased number of cores and faster processing nodes in the new computer cluster will speed up that task so much that Kawai’s graduate student Kyle Bentley can investigate more different cases.
Physicist Kawai describes his computations as the most intensive on campus.“We want to capture information in an image, such as information on an individual’s brain condition,” Skidmore said. “The information we are trying capture, however, can often be difficult to see in the sea of data we collect. One brain may contain millions of bits of data in the form of ‘voxels,’ which are a bit like the pixels on your TV but in three dimensions.”
“When we map to a template,” Skidmore said, “we quadruple the data. When we ask, ‘How does this compare to a healthy brain?’ we double the data again. Then if we look across one brain or across multiple kinds of brain images, the amount of data truly explodes.”
 Physicist Ryoichi Kawai's complex calculations of the electronic structure of a cube of atoms are laying the groundwork for better lasers.“This is made even more complex by the fact that a given image can include more than three dimensions of information,” he said. “One type of image we use generates 5-dimensional brain maps. Since we can’t see in five dimensions, we ask the computer to work in these higher-dimensional spaces to help us pull the information out of the data.”
Physicist Ryoichi Kawai's complex calculations of the electronic structure of a cube of atoms are laying the groundwork for better lasers.“This is made even more complex by the fact that a given image can include more than three dimensions of information,” he said. “One type of image we use generates 5-dimensional brain maps. Since we can’t see in five dimensions, we ask the computer to work in these higher-dimensional spaces to help us pull the information out of the data.”
To look at the adult brain’s plasticity — the ability to change function and structure through new synaptic connections — Visscher studies visual processing.
“Because we look at spatial and temporal data, the number of pieces of information is huge — gigabytes per subject,” she said. “We need to do correlations on all the data points at the same time. To get faster, we optimize the data analysis with a lot of feedback. Then we run what we learned from one brain on a hundred brains.”
The new supercomputer “will drastically improve our computing capacity from what we have had,” Crossman said. “With the old cluster, my job might sit in a queue for a couple of weeks. With undiagnosed diseases, that is not acceptable because that’s a patient.”“And I cannot tell a researcher, ‘I’m sorry, we won’t meet that grant deadline,’” Crossman said. “You know, science can’t stop.”
The data floodgates of genomics burst open about a dozen years ago with the arrival of next-generation, high-throughput sequencing, says Elliot Lefkowitz, Ph.D., director of Informatics for the UAB Center for Clinical and Translational Science. Lefkowitz has been serving the bioinformatics needs of the UAB Center for AIDS Research for 25 years, and now also handles bioinformatics for the UAB Microbiome Facility. His team has grown to five bioinformaticians and several programmers.
“We deal with billions of sequences when we do a run through the DNA sequencing machine,” Lefkowitz said. “We need to compare every one of the billion ‘reads’ (the 50- to 300-base sequence of a short piece of DNA) to every other one. With high-performance computing and thousands of nodes, each one does part of the job.”
“In not too many years,” Lefkowitz speculated, “we will be sequencing every patient coming into University Hospital.”
Changes like that mean ever-increasing computer demands.
“Biomedical research,” Crossman said, “now is big data.”
Computing challenges are found across the UAB campus, from physics and neurology to genetics and the microbiome. Alabama’s most advanced supercomputer is now at UAB, making it possible to solve these challenges.
 Kristina Visscher is using fMRI images from several hundred brains to learn how the brain adapts after long-term changes in vision.What do the human brain, the 3 billion base-pair human genome and a tiny cube of 216 atoms have in common?
Kristina Visscher is using fMRI images from several hundred brains to learn how the brain adapts after long-term changes in vision.What do the human brain, the 3 billion base-pair human genome and a tiny cube of 216 atoms have in common?All of them, from the tiny cube to the 3-pound human brain, create incredibly complex computing challenges for University of Alabama at Birmingham researchers, and aggressive investments in UAB’s IT infrastructure have opened new possibilities in innovation, discovery and patient care.
For example, Kristina Visscher, Ph.D., assistant professor of neurobiology, UAB School of Medicine, uses fMRI images from several hundred human brains to learn how the brain adapts after long-term changes in visual input, such as macular degeneration. Frank Skidmore, M.D., an assistant professor of neurology in the School of Medicine, studies hundreds of brain MRI images to see if they can predict Parkinson’s disease.
David Crossman, Ph.D., bioinformatics director in UAB’s Heflin Center for Genomic Science, deciphers the sequences of human genomes for patients seeking a diagnosis in UAB’s Undiagnosed Diseases Program, and he processes DNA sequencing for UAB researchers who need last-minute data for their research grant applications.
Ryoichi Kawai, Ph.D., an associate professor of physics in the UAB College of Arts and Sciences, is laying the groundwork for a better infrared laser by calculating the electronic structure for a cube made up of just 216 atoms of zinc sulfide doped with chromium or iron.
Each researcher faces a mountain range of computational challenges. Those mountains are now easier to scale with UAB’s new supercomputer — the most advanced in Alabama for speed and memory.
The tiny cube
The 216-atom electronic structure problem “is the largest calculation on campus, by far,” Kawai said. “If you give me 2,000 cores, I can use them. If you give me 10,000 cores, I can use them all without losing efficiency.” (UAB's new supercomputer has 2,304 cores.) Learn more about UAB’s new supercomputer in this story Learn more about UAB’s new supercomputer in this story |
Brains
In their research, both Skidmore and Visscher have to compare brains with other brains. Because each brain differs somewhat in size, shape and surface folds, every brain has to be mapped onto a template to allow comparisons.Physicist Kawai describes his computations as the most intensive on campus.“We want to capture information in an image, such as information on an individual’s brain condition,” Skidmore said. “The information we are trying capture, however, can often be difficult to see in the sea of data we collect. One brain may contain millions of bits of data in the form of ‘voxels,’ which are a bit like the pixels on your TV but in three dimensions.”
“When we map to a template,” Skidmore said, “we quadruple the data. When we ask, ‘How does this compare to a healthy brain?’ we double the data again. Then if we look across one brain or across multiple kinds of brain images, the amount of data truly explodes.”
 Physicist Ryoichi Kawai's complex calculations of the electronic structure of a cube of atoms are laying the groundwork for better lasers.“This is made even more complex by the fact that a given image can include more than three dimensions of information,” he said. “One type of image we use generates 5-dimensional brain maps. Since we can’t see in five dimensions, we ask the computer to work in these higher-dimensional spaces to help us pull the information out of the data.”
Physicist Ryoichi Kawai's complex calculations of the electronic structure of a cube of atoms are laying the groundwork for better lasers.“This is made even more complex by the fact that a given image can include more than three dimensions of information,” he said. “One type of image we use generates 5-dimensional brain maps. Since we can’t see in five dimensions, we ask the computer to work in these higher-dimensional spaces to help us pull the information out of the data.”To look at the adult brain’s plasticity — the ability to change function and structure through new synaptic connections — Visscher studies visual processing.
“Because we look at spatial and temporal data, the number of pieces of information is huge — gigabytes per subject,” she said. “We need to do correlations on all the data points at the same time. To get faster, we optimize the data analysis with a lot of feedback. Then we run what we learned from one brain on a hundred brains.”
UAB customers
Crossman deciphers the sequences of human genomes for patients seeking a diagnosis, and he processes DNA sequencing for UAB researchers. Providing excellent customer service to his clients is vital, Crossman says, and it takes computer power to crunch genome sequencing data for those researchers, physicians and patients.
Managing traffic on the new UAB computer clusterSize:
|
The data floodgates of genomics burst open about a dozen years ago with the arrival of next-generation, high-throughput sequencing, says Elliot Lefkowitz, Ph.D., director of Informatics for the UAB Center for Clinical and Translational Science. Lefkowitz has been serving the bioinformatics needs of the UAB Center for AIDS Research for 25 years, and now also handles bioinformatics for the UAB Microbiome Facility. His team has grown to five bioinformaticians and several programmers.
“We deal with billions of sequences when we do a run through the DNA sequencing machine,” Lefkowitz said. “We need to compare every one of the billion ‘reads’ (the 50- to 300-base sequence of a short piece of DNA) to every other one. With high-performance computing and thousands of nodes, each one does part of the job.”
“In not too many years,” Lefkowitz speculated, “we will be sequencing every patient coming into University Hospital.”
Changes like that mean ever-increasing computer demands.
“Biomedical research,” Crossman said, “now is big data.”
McKnight Poster Session
By: Vicki Hixon
Published Date: Aug 12
Hosted by:
Evelyn F. McKnight Brain Institute, The University of Alabama at Birmingham
Evelyn F. McKnight Brain Institute, The University of Arizona
Evelyn F. & William L. McKnight Brain Institute, The University of Florida
Evelyn F. McKnight Brain Institute, The University of Miami
Sponsored by:
McKnight Brain Research Foundation
Established by Evelyn F. McKnight
You are cordially invited to the
Ninth Annual
McKnight Brain Research Foundation
Poster Reception
Sunday, November 13, 2016
6:30 - 8:30 p.m.
Hilton San Diego Bayfront
One Park Boulevard
San Diego, CA 92101
Hosted by:
Evelyn F. McKnight Brain Institute, The University of Alabama at Birmingham
Evelyn F. McKnight Brain Institute, The University of Arizona
Evelyn F. & William L. McKnight Brain Institute, The University of Florida
Evelyn F. McKnight Brain Institute, The University of Miami
Sponsored by:
McKnight Brain Research Foundation
Established by Evelyn F. McKnight
Art Wanted
By: Vicki Hixon
Published Date: Aug 04


Discovery may lead to a treatment to slow Parkinson's disease
By: Vicki Hixon
Published Date: Jul 27
Discovery may lead to a treatment to slow Parkinson’s disease by Jeff Hansen
Researchers have found that an interaction between a mutant gene and alpha synuclein in neurons leads to hallmark pathologies seen in Parkinson’s disease, findings that may lead to new mechanisms and targets for neuroprotection.
 Laura A. Volpicelli-DaleyUsing a robust model for Parkinson’s disease, University of Alabama at Birmingham researchers and colleagues have discovered an interaction in neurons that contributes to Parkinson’s disease, and they have shown that drugs now under development may block the process.
Laura A. Volpicelli-DaleyUsing a robust model for Parkinson’s disease, University of Alabama at Birmingham researchers and colleagues have discovered an interaction in neurons that contributes to Parkinson’s disease, and they have shown that drugs now under development may block the process.
The research team has shown that the most common genetic cause of Parkinson’s disease — a mutant LRRK2 kinase enzyme — contributes to the formation of inclusions in neurons, resembling one of the hallmark pathologies seen in Parkinson’s disease. These inclusions are made up of aggregated alpha synuclein protein, which — the research also shows — can be prevented from forming by using two LRRK2 kinase inhibitor drugs now being developed for clinical use.
The interaction between mutant LRRK2 kinase and alpha-synuclein “may uncover new mechanisms and targets for neuroprotection,” the researchers write in a recent Journal of Neuroscience paper. “These results demonstrate that alpha-synuclein inclusion formation in neurons can be blocked and that novel therapeutic compounds targeting this process by inhibiting LRRK2 kinase activity may slow progression of Parkinson’s disease-associated pathology.”
The potential clinical applications for novel neuroprotection strategies in LRRK2-linked Parkinson’s need to be tested in other preclinical models of Parkinson’s disease, say the researchers, led by corresponding author Laura A. Volpicelli-Daley, Ph.D., and senior author Andrew B. West, Ph.D., Center for Neurodegeneration and Experimental Therapeutics, UAB Department of Neurology.
“These data give us hope for the clinical potential of LRRK2 kinase inhibitors as effective therapies for Parkinson’s disease,” Volpicelli-Daley said. “The LRRK2 kinase inhibitors may inhibit the spread of pathologic alpha-synuclein, not only in patients with LRRK2 mutations, but in all Parkinson’s disease patients. Future studies to validate the safety and efficacy of the LRRK2 inhibitors will be necessary before testing the inhibitors in human clinical trials.”
Besides Parkinson’s disease, alpha-synuclein also plays a central role in development of dementia with Lewy bodies and multiple system atrophy, and it is associated with Alzheimer’s disease and other neurodegenerative disorders.
The Parkinson’s disease model developed by Volpicelli-Daley applies very low concentrations of pre-formed fibrils of alpha-synuclein to in vitro or in vivo neurons. This causes formation of modified alpha-synuclein inclusions that share morphology with those found in the Parkinson’s disease brain after death.
They used this model to test the effects of neuron expression of the mutant LRRK2 (“lark two”) kinase, G2019S-LRRK2, on the formation of the inclusion pathology.
They found that:
 Andrew B. WestIn fluorescence-recovery-after-photobleaching experiments, they found there was a larger pool of mobile alpha-synuclein, as opposed to membrane-bound alpha-synuclein, in neurons that expressed G2019S-LRRK2. Recent work by others has shown that mobile alpha-synuclein is prone to misfolding and aggregation, so the researchers hypothesize that the G2019S-LRRK2 mutation may contribute to Parkinson’s susceptibility by boosting the amounts of mobile alpha-synuclein in neurons.
Andrew B. WestIn fluorescence-recovery-after-photobleaching experiments, they found there was a larger pool of mobile alpha-synuclein, as opposed to membrane-bound alpha-synuclein, in neurons that expressed G2019S-LRRK2. Recent work by others has shown that mobile alpha-synuclein is prone to misfolding and aggregation, so the researchers hypothesize that the G2019S-LRRK2 mutation may contribute to Parkinson’s susceptibility by boosting the amounts of mobile alpha-synuclein in neurons.
Besides Volpicelli-Daley and West, co-authors of the paper “G2019S-LRRK2 expression augments alpha-synuclein sequestration into inclusions in neurons” are Hisham Abdelmotilib, Zhiyong Liu, Lindsay Stoyka, João Paulo Lima Daher and Kyle Fraser, all of the Center for Neurodegeneration and Experimental Therapeutics, UAB Department of Neurology; Austen J. Milnerwood, Centre for Applied Neurogenetics, University of British Columbia; Vivek K. Unni, Jungers Center for Neurosciences Research and Parkinson Center of Oregon, Oregon Health & Science University; Warren D. Hirst, Pfizer Neuroscience and Pain Research Unit, Cambridge, Massachusetts; Zhenyu Yue, Departments of Neurology and Neuroscience, Icahn School of Medicine at Mount Sinai; Hien T. Zhao, Ionis Pharmaceuticals, Carlsbad, California; and Richard E. Kennedy, Comprehensive Center for Healthy Aging and Division of Gerontology, Geriatrics, and Palliative Care, UAB Department of Medicine.
Grants to fund this work came from the American Parkinson’s Disease Association, the Michael J. Fox Foundation LEAPS Award and the National Institutes of Health NS064934.
Volpicelli-Daley is an assistant professor in the Department of Neurology.
West is co-director of the Center for Neurodegeneration and Experimental Therapeutics, and the John A. and Ruth R. Jurenko Professor of Neurology at UAB.
 Laura A. Volpicelli-DaleyUsing a robust model for Parkinson’s disease, University of Alabama at Birmingham researchers and colleagues have discovered an interaction in neurons that contributes to Parkinson’s disease, and they have shown that drugs now under development may block the process.
Laura A. Volpicelli-DaleyUsing a robust model for Parkinson’s disease, University of Alabama at Birmingham researchers and colleagues have discovered an interaction in neurons that contributes to Parkinson’s disease, and they have shown that drugs now under development may block the process.The research team has shown that the most common genetic cause of Parkinson’s disease — a mutant LRRK2 kinase enzyme — contributes to the formation of inclusions in neurons, resembling one of the hallmark pathologies seen in Parkinson’s disease. These inclusions are made up of aggregated alpha synuclein protein, which — the research also shows — can be prevented from forming by using two LRRK2 kinase inhibitor drugs now being developed for clinical use.
The interaction between mutant LRRK2 kinase and alpha-synuclein “may uncover new mechanisms and targets for neuroprotection,” the researchers write in a recent Journal of Neuroscience paper. “These results demonstrate that alpha-synuclein inclusion formation in neurons can be blocked and that novel therapeutic compounds targeting this process by inhibiting LRRK2 kinase activity may slow progression of Parkinson’s disease-associated pathology.”
The potential clinical applications for novel neuroprotection strategies in LRRK2-linked Parkinson’s need to be tested in other preclinical models of Parkinson’s disease, say the researchers, led by corresponding author Laura A. Volpicelli-Daley, Ph.D., and senior author Andrew B. West, Ph.D., Center for Neurodegeneration and Experimental Therapeutics, UAB Department of Neurology.
“These data give us hope for the clinical potential of LRRK2 kinase inhibitors as effective therapies for Parkinson’s disease,” Volpicelli-Daley said. “The LRRK2 kinase inhibitors may inhibit the spread of pathologic alpha-synuclein, not only in patients with LRRK2 mutations, but in all Parkinson’s disease patients. Future studies to validate the safety and efficacy of the LRRK2 inhibitors will be necessary before testing the inhibitors in human clinical trials.”
Besides Parkinson’s disease, alpha-synuclein also plays a central role in development of dementia with Lewy bodies and multiple system atrophy, and it is associated with Alzheimer’s disease and other neurodegenerative disorders.
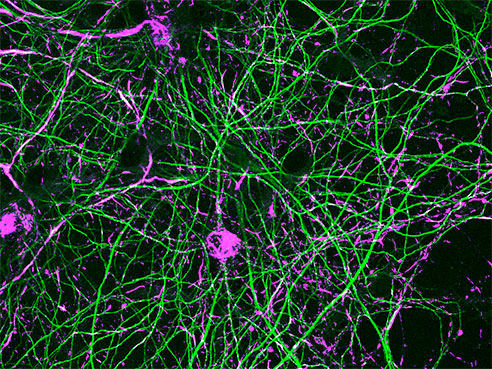 Primary hippocampal neurons from mice expressing G2019S-LRRK2. The neurons were treated with alpha-synuclein fibrils, and 18 days later immunofluorescence was performed. The magenta shows phospho-alpha-synuclein inclusions in the cell bodies and throughout the axons, which are visualized as green.Research details
Primary hippocampal neurons from mice expressing G2019S-LRRK2. The neurons were treated with alpha-synuclein fibrils, and 18 days later immunofluorescence was performed. The magenta shows phospho-alpha-synuclein inclusions in the cell bodies and throughout the axons, which are visualized as green.Research details
The Parkinson’s disease model developed by Volpicelli-Daley applies very low concentrations of pre-formed fibrils of alpha-synuclein to in vitro or in vivo neurons. This causes formation of modified alpha-synuclein inclusions that share morphology with those found in the Parkinson’s disease brain after death.They used this model to test the effects of neuron expression of the mutant LRRK2 (“lark two”) kinase, G2019S-LRRK2, on the formation of the inclusion pathology.
They found that:
- G2019S-LRRK2 enhanced alpha-synuclein inclusions in primary hippocampal neurons from the hippocampus region of the brain, 18 days after fibril exposure, as compared with neurons that over-expressed normal LRRK2.
- The effects of G2019S-LRRK2 expression in the fibril-exposed neurons were lessened by very low concentrations of potent and selective preclinical drugs that inhibit LRRK2 kinase. This suggested that the kinase activity of G2019S-LRRK2, which adds a phosphate onto target proteins, underlies the faster formation of pathologic alpha-synuclein inclusions.
- G2019S-LRRK2 expression enhanced alpha-synuclein inclusion formation in dopamine neurons from the region of the brain called the substantia nigra pars compacta. The substantia nigra pars compacta is the area of the brain that dies in Parkinson’s disease, so this experiment further supports a link between the G2019S-LRRK2 mutation and Parkinson’s pathogenesis.
 Andrew B. WestIn fluorescence-recovery-after-photobleaching experiments, they found there was a larger pool of mobile alpha-synuclein, as opposed to membrane-bound alpha-synuclein, in neurons that expressed G2019S-LRRK2. Recent work by others has shown that mobile alpha-synuclein is prone to misfolding and aggregation, so the researchers hypothesize that the G2019S-LRRK2 mutation may contribute to Parkinson’s susceptibility by boosting the amounts of mobile alpha-synuclein in neurons.
Andrew B. WestIn fluorescence-recovery-after-photobleaching experiments, they found there was a larger pool of mobile alpha-synuclein, as opposed to membrane-bound alpha-synuclein, in neurons that expressed G2019S-LRRK2. Recent work by others has shown that mobile alpha-synuclein is prone to misfolding and aggregation, so the researchers hypothesize that the G2019S-LRRK2 mutation may contribute to Parkinson’s susceptibility by boosting the amounts of mobile alpha-synuclein in neurons.Besides Volpicelli-Daley and West, co-authors of the paper “G2019S-LRRK2 expression augments alpha-synuclein sequestration into inclusions in neurons” are Hisham Abdelmotilib, Zhiyong Liu, Lindsay Stoyka, João Paulo Lima Daher and Kyle Fraser, all of the Center for Neurodegeneration and Experimental Therapeutics, UAB Department of Neurology; Austen J. Milnerwood, Centre for Applied Neurogenetics, University of British Columbia; Vivek K. Unni, Jungers Center for Neurosciences Research and Parkinson Center of Oregon, Oregon Health & Science University; Warren D. Hirst, Pfizer Neuroscience and Pain Research Unit, Cambridge, Massachusetts; Zhenyu Yue, Departments of Neurology and Neuroscience, Icahn School of Medicine at Mount Sinai; Hien T. Zhao, Ionis Pharmaceuticals, Carlsbad, California; and Richard E. Kennedy, Comprehensive Center for Healthy Aging and Division of Gerontology, Geriatrics, and Palliative Care, UAB Department of Medicine.
Grants to fund this work came from the American Parkinson’s Disease Association, the Michael J. Fox Foundation LEAPS Award and the National Institutes of Health NS064934.
Volpicelli-Daley is an assistant professor in the Department of Neurology.
West is co-director of the Center for Neurodegeneration and Experimental Therapeutics, and the John A. and Ruth R. Jurenko Professor of Neurology at UAB.
Research Grant Proposals
By: Vicki Hixon
Published Date: Jul 25
The Civitan International Research Center Research Grants for Emerging Scholars Request for Proposals
Civitan Travel Awards
Day Published in Nature Communications
By: Vicki Hixon
Published Date: Jul 07
Extra-coding RNAs regulate DNA methylation in the adult brain
Pozzo-Miller Lab and Dudek Lab Collaboration
By: Kerry Gorelick
Published Date: Jun 23
Perineuronal Nets Suppress Plasticity of Excitatory Synapses on CA2 Pyramidal Neurons
Kelly E. Carstens, Mary L. Phillips, Lucas Pozzo-Miller, Richard J. Weinberg, and Serena M. Dudek
The Journal of Neuroscience, 8 June 2016, 36(23):6312-6320; doi:10.1523/JNEUROSCI.0245-16.2016 FEATURED ARTICLE COVER ILLUSTRATION
http://www.jneurosci.org/content/36/23/6312.abstract
Kelly E. Carstens, Mary L. Phillips, Lucas Pozzo-Miller, Richard J. Weinberg, and Serena M. Dudek
The Journal of Neuroscience, 8 June 2016, 36(23):6312-6320; doi:10.1523/JNEUROSCI.0245-16.2016 FEATURED ARTICLE COVER ILLUSTRATION
http://www.jneurosci.org/content/36/23/6312.abstract
 Perineuronal Nets Suppress Plasticity of Excitatory Synapses on CA2 Pyramidal Neurons
Perineuronal Nets Suppress Plasticity of Excitatory Synapses on CA2 Pyramidal Neurons