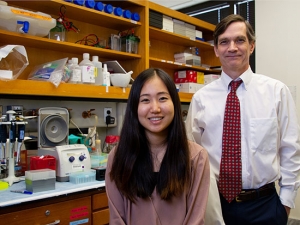
Assistant Professor Shahid Mukhtar, Ph.D., was part of the first research team to build an Arabidopsis immune interactome map — a map of the tens of thousands of interactions that link proteins from pathogens and host and host immune proteins with other host proteins.
 Assistant Professor Shahid Mukhtar, right, was part of the first research team to build a plant interactome map. |
Now he hopes to be part of a team that will develop an interactome map for humans.
Mukhtar is working with Robert Kimberly, M.D., senior associate dean for research in the School of Medicine on the project. The success Mukhtar had in building the plant immune interactome gave him clues into ways pathogens can hijack cellular machinery to cause disease — and the confidence that the same sort of model can be built for humans.
“Human innate immunity forms the first and perhaps the most critical line of defense against invading pathogens,” Mukhtar says. “Any disorder in the immune system can result in disease, including autoimmune diseases, inflammatory diseases and cancer.
“It would be a tremendous achievement if we can find a way to reveal how human immune receptors can activate immune responses.”
Mukhtar and Kimberly will collaborate with scientists with the Dana Farber Cancer Institute in Boston, Mass.
The years of work building the plant immune interactome gives Mukhtar confidence the same can be achieved in humans. The findings from the plant interactome project were published in the July 29, 2011, issue of the journal Science and suggest that while pathogens employ a diverse arsenal of weapons, they attack only a limited number of cellular targets.
The study mapped one-third of the interactome of proteins encoded by the genome of the plant Arabidopsis thaliana, or thale cress, a model plant. Arabidopsis has traits that make it useful for understanding the immune mechanisms in other plant species.
“Just to get where we are today we did robotic screening of millions of combinations — at least more than 12 million,” says Mukhtar, who came to UAB from the University of North Carolina at Chapel Hill in fall 2010. “In all that work, during the course of five years, I never got frustrated or lost hope because you could see an interactome map developing that may provide a path to a life-changing discovery.”
Kimberly says the success of plant interactome map shows the value of model organisms to science and gives hope to those fighting autoimmune diseases.
“The success of this study shows how valuable model organisms can be in understanding fundamental principles in biology,” says Kimberly. “We are now looking to build a human interactome at UAB in our continuing efforts to fight diseases such as arthritis, diabetes, Crohn’s and cancer.”
Mukhtar’s goal is to work with Kimberly to build the human interactome map using at lest 16,000 human proteins to see how the receptors interact with other proteins in humans and how the different virulence proteins from the pathogens attack on some proteins in humans.
“In order to fight back with a pathogen, we need to find out where they attack inside the host cell,” Mukhtar says. “If we wanted to do genetic or chemical manipulation to interfere with those types of interactions, we need to first fine out where the pathogen attacks the human.”
Biofuel project
Mukhtar also will continue work on another research project of significance — the creation of biofuel using algae.
Biofuel production is an inefficient process. Currently, biofuels derived from corn and other feed crops reduce the amount of arable land that can be used for other important purposes. Mukhtar says using biofuels derived from algae is potentially more efficient and has a much lesser impact on the environment.
“Microalgae can grow rapidly and produce more fuel on a per-acre basis than other sources of biofuels,” Mukhtar says. “Algae thrive in non-arable areas of both marine and freshwater. In this way, algae can be used in areas where other organisms would not survive. In fact, algae can use higher concentrations of carbon dioxide in order to grow. Mass algae culturing for biofuel also would serve to mitigate higher levels of green-house gases around industrial areas.”
Despite the advantages algae offers as a biofuel source, producing it in mass quantities has proven inefficient.
Mukhtar wants to improve the process in his lab. He is doing so by creating mutant algae strains that overproduce lipids, which is the source of biofuel.
“To achieve this, we use flow cytometry cell-sorting and gravimetric isolation of lipids,” he says. “Additionally, we study the lipid pathway at the gene level using cutting-edge technology such as real-time PCR, next-generation sequencing and comparative transcriptomics. This will lead to an improvement in the use of algae as biofuel in the near future.”