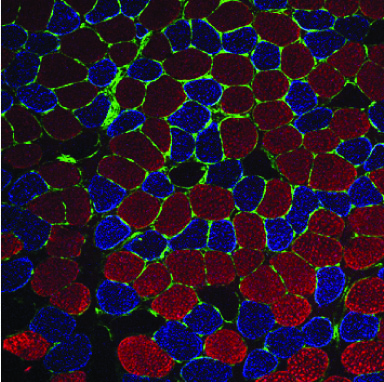
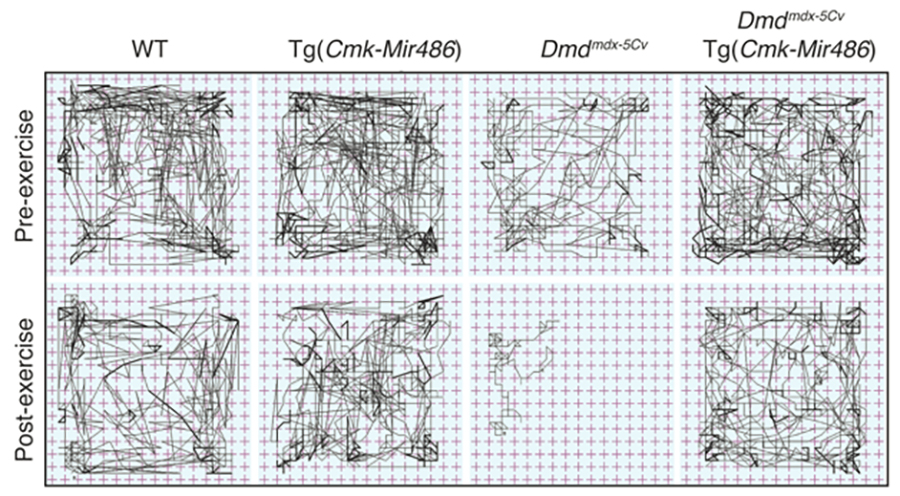
 Non-coding RNAs as modifiers of neuromuscular disease- The Alexander lab is especially interested in determining the functional roles of non-coding RNAs (microRNA, lncRNAs, and snRNAs) in both normal and diseased muscle. We identified one particular microRNA, miR-486, as a muscle-enriched miRNA that is strongly reduced in expression levels in Duchenne muscular dystrophy (DMD), but not the milder Becker muscular dystrophy (BMD). Additionally, we demonstrated that miR-486 overexpression in dystrophic mouse muscles can block the progression of DMD symptoms and improve overall muscle performance and physiology (Alexander et al., JCI, 2014). We are now characterizing the targets of miR-486 in skeletal muscle, in addition to determining if miR-486 expression levels can be used as a biomarker for disease progression in DMD and other muscular dystrophies.
Non-coding RNAs as modifiers of neuromuscular disease- The Alexander lab is especially interested in determining the functional roles of non-coding RNAs (microRNA, lncRNAs, and snRNAs) in both normal and diseased muscle. We identified one particular microRNA, miR-486, as a muscle-enriched miRNA that is strongly reduced in expression levels in Duchenne muscular dystrophy (DMD), but not the milder Becker muscular dystrophy (BMD). Additionally, we demonstrated that miR-486 overexpression in dystrophic mouse muscles can block the progression of DMD symptoms and improve overall muscle performance and physiology (Alexander et al., JCI, 2014). We are now characterizing the targets of miR-486 in skeletal muscle, in addition to determining if miR-486 expression levels can be used as a biomarker for disease progression in DMD and other muscular dystrophies. Epigenetic regulation of normal and diseased muscle- We are also very interested in uncovering novel epigenetic regulators of normal and diseased skeletal muscle. We seek to answer the questions such as how can two genetically-related brothers with the exact same pathogenic disease mutation have two dramatically different clinical phenotypes? Our past work uncovered the JAG1 genomic locus as a modifier of DMD (Vieira et al., Cell, 2015). We are actively seeking to identify additional loci that modify the disease pathology and/or progression in DMD and other muscular dystrophies. We are currently characterizing several histone modifier and RNA-splicing proteins that may affect muscle formation and maintenance in our vertebrate animal models.
Epigenetic regulation of normal and diseased muscle- We are also very interested in uncovering novel epigenetic regulators of normal and diseased skeletal muscle. We seek to answer the questions such as how can two genetically-related brothers with the exact same pathogenic disease mutation have two dramatically different clinical phenotypes? Our past work uncovered the JAG1 genomic locus as a modifier of DMD (Vieira et al., Cell, 2015). We are actively seeking to identify additional loci that modify the disease pathology and/or progression in DMD and other muscular dystrophies. We are currently characterizing several histone modifier and RNA-splicing proteins that may affect muscle formation and maintenance in our vertebrate animal models.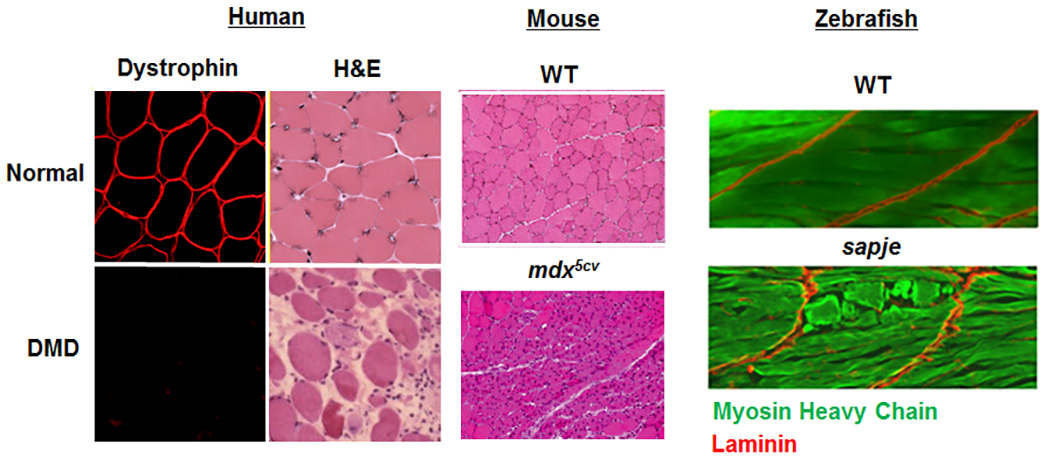
{slide=Publications}
- Vieira NM, Elvers I, Alexander MS, Moreira YB, Eran A, Gomes JP, Marshall JL, Karlsson EK, Verjovski-Almeida S, Lindblad-Toh K, Kunkel LM, and Zatz M. Jagged 1 rescues the Duchenne muscular dystrophy phenotype. Cell. 2015. 163(5):1204-1213. PMCID: 4668935.
- Alexander MS, Casar JC, Motohashi N, Vieira NM, Eisenberg I, Marshall JL, Gasperini MJ, Lek A, Myers JA, Estrella EA, Kang PB, Shapiro F, Rahimov F, Kawahara G, Widrick JJ, Kunkel LM. MicroRNA-486-dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy-associated symptoms. J. Clinical Investigation. 2014. 124(6):2651-2667. PMCID: 4038577.
- Alexander MS, Kawahara G, Motohashi N, Casar JC, Eisenberg I, Myers JA, Gasperini MJ, Estrella EA, Kho AT, Mitsuhashi S, Shapiro F, Kang PB, Kunkel LM. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death and Differentiation. 2013. 20(9):1194-1208. PMCID: 3741500.
- Motohashi N*, Alexander MS*, Shimizu-Motohashi Y, Myers JA, Kawahara G, Kunkel LM. Regulation of IRS1/Akt insulin signaling by microRNA-128a during myogenesis. J. Cell Science. 2013. 126(Pt.12):2678-91. PMCID: 3687700.
- Alexander MS, Casar JC, Motohashi N, Myers JA, Eisenberg I, Gonzalez RT, Estrella EA, Kang PB, Kawahara G, Kunkel LM. Regulation of DMD pathology by an ankyrin-encoded microRNA. Skelet. Musc. 2011;1(27). PMCID: 3188430.
- Kawahara G, Karpf JA, Myers JA, Alexander MS, Guyon, JR, Kunkel LM. Drug screening in a zebrafish model of Duchenne Muscular Dystrophy. Proc. Natl. Acad. Sci. 2011. 108(13):5331-5336. PMCID: 3069215.
- Alexander MS, Kawahara G, Kho AT, Howell MH, Pusack TJ, Myers JA, Montanaro F, Zon LI, Guyon JR, and Kunkel LM. Isolation and transcriptome analysis of adult zebrafish cell enriched for skeletal muscle progenitors. Muscle and Nerve 2011 43(5):741-50. PMCID: 3075361.
- Alexander MS, Shi X, Voelker KA, Grange RW, Garcia JA, Hammer RE, Garry DJ. Foxj3 transcriptionally activates Mef2c and regulates adult skeletal muscle fiber type identity. Dev. Biol. 2010; 337(2):396-404. PMCID: Not Federally Funded.
- Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science 2009; 324(5932):1289-1293. Not Federally Funded.
- Meeson, AP*, Shi X*, Alexander MS*, Williams RS, Allen RE, Jiang N, Adham IM, Goetsch SC, Hammer RE, Garry DJ. Sox15 and Fhl3 transcriptionally coactivate Foxk1 and regulate myogenic progenitor cells. EMBO J. 2007; 26(7):1902-1912. PMCID: PMC1847663.
{/slide}








{/slide}
Opportunities
The Alexander lab is activity seeking to add individuals to join an exciting team of scientists. We are actively seeking a Research Technician and Postdoctoral Fellow. Technical skills in molecular biology, biochemistry, cell biology, and mouse/zebrafish husbandry are highly desired. For inquiries about the Research Technician or Postdoctoral fellow positions, please contact Dr. Alexander directly via email along with sending your CV and the names/contact information for at least 2 references.
Undergraduate students interested in summer rotations should contact Dr. Alexander directly via email. Undergraduates with computational biology backgrounds interested in research should contact Dr. Alexander directly via email.
