
-
Mining the Genome - Issue 15 / Undiagnosed Diseases Program Joins the NIH National Network
The Undiagnosed Diseases Program (UDP) is a clinical program that launched at the University of Alabama at Birmingham in 2013. The program is funded by institutional support and revenue generated from patient visits. The UDP aims to secure a diagnosis for patients with rare conditions or unusual presentations of not-so-rare conditions, and in the process, to advance medical knowledge about disease. The UDP team has been successful in assigning a diagnosis to upwards of 300 patients, each of whom has arrived at the program after already searching for a diagnosis for years. Now the program has been selected as an official affiliate site in the NIH National Undiagnosed Disease Network, meaning more connection to other program sites, national resources, and opportunities to work with more patients from across the country. This new status may also present a new funding mechanism for the Program in the future.
Dr. Bruce Korf, director of CCTS Genomic Medicine and Associate Dean for Genomic Medicine, UAB School of Medicine, Chief Genomics Officer, UAB Medicine, along with other members of the UAB UDP team, is pleased to also share collective knowledge across the UDP network, learning from the other affiliate sites and presenting what his team has learned as well. Learn more about the Program and team by reading the original UDP Mining the Genome post below or by clicking here. -
Mining the Genome - Issue 14 / Implementing Genomics in Practice (IGNITE)
In December of last year, Nita Limdi, PharmD, PhD, MSPH, FAHA, received an invitation to lead a new clinical research site for Implementing Genomics in Practice (IGNITE), a National Human Genome Research Institute (NHGRI) project intended to stimulate research on genomic medicine. Dr. Limdi’s longstanding track record in Pharmacogenomics and her sustained contributions to the field set the stage for this opportunity. This exciting invitation makes the CCTS Hub (UAB) a participant in three key NHGRI programs (IGNITE, eMERGE, and CSER).
IGNITE is centered on genomic medicine research, with the goal to improve patient health and treatments and to better predict a patient’s risk for disease. The project consists of two major clinical trial studies at this time. UAB joins the first, pursuing “Genetic Testing to Understand and Address Renal Disease Disparities in the United States”. The second is titled “A Depression and Opioid Pragmatic Trial in Pharmacogenetics” and is being conducted at several other sites across the United States. IGNITE is being led by seven investigators across six institutions. The sites are working together to develop new methods to incorporate genomic information into the electronic medical record and to provide clinicians support for what to do with that information. Then the sites will disseminate their findings so that new methods and best practices can be shared.
Dr. Limdi and her team begin recruitment for IGNITE in September 2022. Stay tuned to the Digest and Mining the Genome for updates to their progress and results.
-
Mining the Genome - Issue 13 / All of Us Research Program Data Funding Opportunity
Eligible grantees of participating Institutes, Centers, and Offices are encouraged to apply for up to $75,000 in direct costs to support analysis of currently available data within the All of Us Research Program’s Researcher Workbench or to develop analysis tools for the available data within the general scope of the parent award. Proposals that advance precision medicine and address COVID-19 exposure, treatment and Post-Acute Sequelae of SARS CoV-2 (long COVID) are encouraged.
The All of Us data includes nearly 100,000 highly diverse whole genome sequences are available and about 50% of the data is from individuals who identify with racial or ethnic groups that have historically been underrepresented in research.
Deadline to apply: July 5, 2022. Learn more here.
-
Mining the Genome - Issue 12 / All of Us Research Program Releases Nearly 100,000 Whole Genome Sequences through the Researcher Workbench
Nearly 100,000 highly diverse whole genome sequences are now available through the NIH’s All of Us Research Program. About 50% of the data is from individuals who identify with racial or ethnic groups that have historically been underrepresented in research. For about a year researchers have had access to other data points from All of Us, but this release enables genome and phenome-wide association studies, as well as other genomic analyses.
These datasets will enable researchers to address yet unanswered questions about health and disease, leading to new breakthroughs and advancing discoveries to reduce persistent health disparities. The genomic data is available via a cloud-based platform, the All of Us Researcher Workbench, and also includes genotyping arrays from 165,000 participants. Whole genome sequencing provides information about almost all of an individual’s genetic makeup, while genotyping arrays capture a specific subset of the genome.
"This release allows an exciting opportunity for researchers to examine genetic factors that contribute to health and disease using the unprecedentedly large and diverse All of Us dataset. After all the effort at engagement, recruitment, and retention in the program, this is a major step forward towards realizing the scientific payoff from this historic project," shares Dr. Bruce Korf, director of CCTS Genomic Medicine.
Get started by visiting researchallofus.org and completing the required registration. Once you are granted access you can begin to review the data, assemble cohorts and create queries. While the data is not downloadable, the Researcher Workbench provides effective and powerful tools for calculations on the spot. There are several video tutorials available to help new users get started.
Contact the CCTS if you have questions about getting started in the All of Us Researcher Workbench. -
Mining the Genome - Issue 11 / Check Out the All of Us Research Program
The All of Us Research Program aims to enroll a million people across the country to build a large health database that will help researchers learn how our biology, lifestyle, and environment affect health. Participants provide blood and urine samples and body measurements and agree to share their electronic health records. Through questionnaires, they provide data points like activity level and environmental exposures. Data from fitness trackers may also be used in some instances.
Participants will also have their genomes sequenced, resulting in even richer research possibilities next year. In the meantime, discovery the many wonderful opportunities available now:
- Diversity is why this population data stands out. All of Us is focused on enrolling individuals from groups historically underrepresented in biomedical research, resulting in the most diverse biomedical dataset ever assembled. Black and African Americans, and Hispanic and Latinx groups are robustly represented, along with individuals from disadvantaged backgrounds.
- Access is simple. The hub includes different tiers of access, with the first tier being available to anyone who visits the website, providing aggregated data. The next tier provides access to a Researcher Workbench, with more detailed, yet anonymized, data and requires registration through an institution already established on the site, but if your institution is not yet represented, the process isn’t burdensome. After completing some forms and simple training, you can begin to identify and create cohorts. To note while exploring data at this level:
- You cannot download the data. The Researcher Workbench, provides various that you can use to perform data analysis, along with the option to request assistance with other analysis needs.
- IRB approval is generally not required to view data and create cohorts (remembering that IRB rules may differ across institutions, so ensure compliance on a case by case basis).
- Researchers who tap into the hub are currently given $300 credit for computational data services, sufficient to conduct meaningful analyses currently.
- Training opportunities are continuous, with video assets already in existence on the hub website and more to come.
- Whole genome sequencing data is coming. Within the next year or so, the All of Us Research Hub will contain participants’ whole genome sequencing, enabling genome- and phenome-wide association studies, as well as other genomic analyses. Access to genomic data may require additional researcher training and certification.
Get set up in the All of Us Research Hub today by visiting researchallofus.org. Please feel free to address questions by contacting CCTS Director of Genomic Medicine,
This email address is being protected from spambots. You need JavaScript enabled to view it. . -
Mining the Genome - Issue 10 / How the Undiagnosed Diseases Program Can Help Researchers & Trainees Across the Partner Network
The Undiagnosed Diseases Program (UDP) is a clinical program that launched at the CCTS Hub (UAB) in 2013. The UDP team aims to find a diagnosis for patients with rare conditions or unusual presentations of not-so-rare conditions, and in the process, to advance medical knowledge about disease. The UDP team has been successful in assigning a diagnosis to approximately half of the patients they’ve evaluated thus far, a significant feat considering the fact that every patient’s case has already been thoroughly evaluated prior to entering the UDP program. (For more detailed information about the process behind the program, scroll down the page to Mining the Genome Issue 5).
For people who have been on a diagnostic odyssey, the program’s potential benefits are clear. But the UDP also provides meaningful opportunity for researchers and trainees across the CCTS Partner Network. Here are three ways you may benefit from involvement in the program:
-
Participate in patient case conferences. All are welcome to join these monthly meetings where clinicians from every specialty gather around the virtual table to discuss clinical cases in the effort to achieve a diagnosis. Contact nurse practitioner
This email address is being protected from spambots. You need JavaScript enabled to view it. for more information on how to join.
-
Refer patients. Patients from across all of the CCTS Partner sites are eligible for evaluation by the UDP. Furthermore, Partners can explore the potential in creating local nodes of the same type of program, replicating some of the procedures that have been successful at the Hub and putting in place a team to help patients locally. Contact CCTS Genomic Medicine director
This email address is being protected from spambots. You need JavaScript enabled to view it. , to learn more.
-
Enhance your research. UDP frequently generates research opportunities. When the UDP team makes a diagnosis and identifies a genetic cause, it could open doors for a researcher studying a certain mutation, gene or cellular process. Their research could now include the knowledge gleaned from a patient in the program. The UDP team will soon make available to the Partner Network a list of the diagnoses made thus far, along with the genetic findings from each. There is also an excellent opportunity to study the diagnostic process itself. Now that there are a few hundred UDP diagnoses completed, there is an opportunity to look back over the course of the program and identify any clues that could have allowed the team to reach a conclusion sooner. Furthermore, incorporating machine learning and artificial intelligence would allow a researcher to fully analyze the successes in order to yield more success for patients in the future.
The UDP has been an invaluable resource to patients who have been seeking a diagnosis, often for many years, but it has also been a resource for both training and research. The UAB UDP team welcomes involvement from CCTS Partners to share in these benefits and enhance the effectiveness of the program.
This newly published CCTS poster explores the collaborative path from diagnostic odysseys to drug discoveries.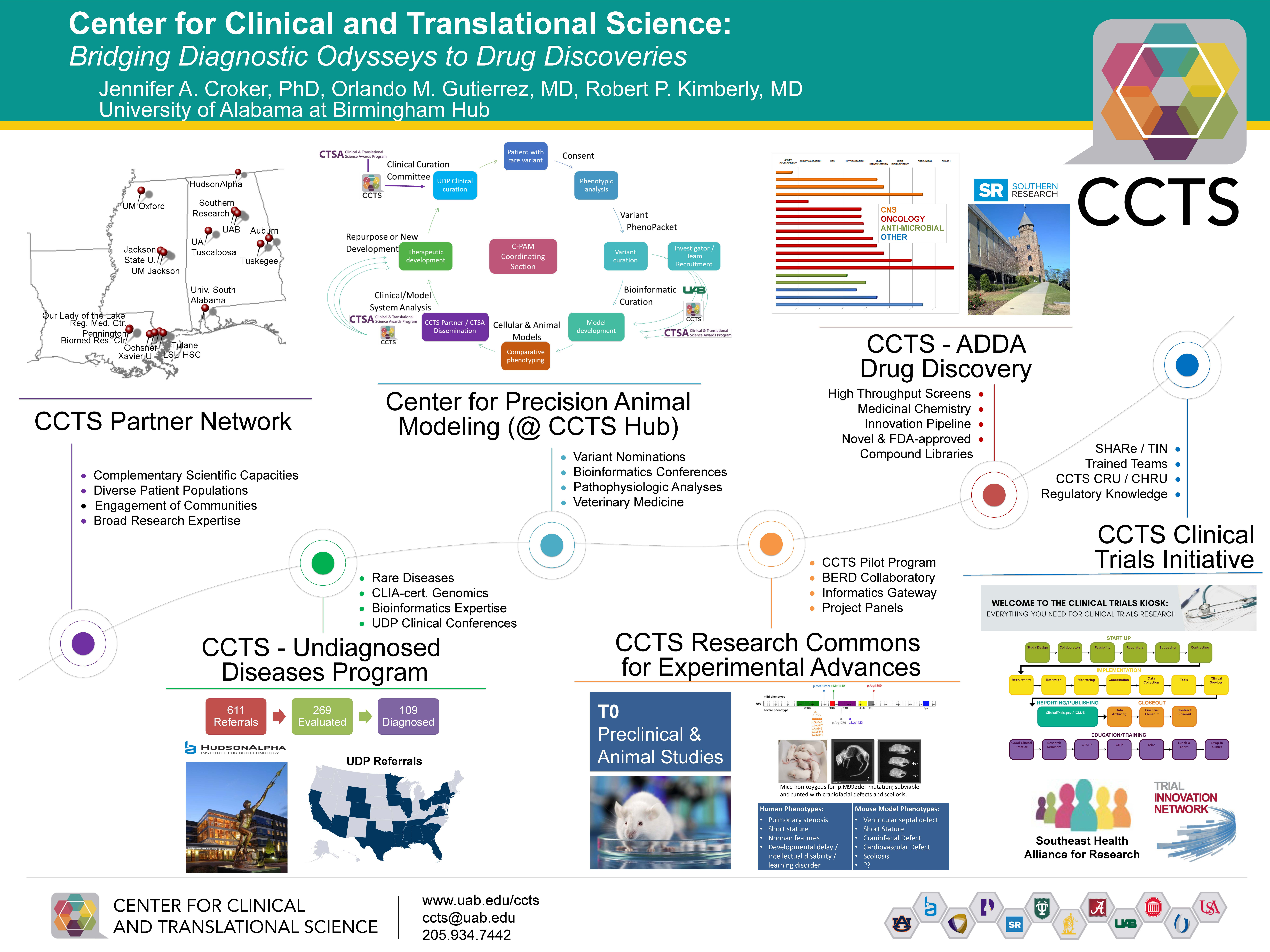 View the brief poster presentation here.
View the brief poster presentation here. -
-
Mining the Genome - Issue 9 / Newborn Genome Sequencing
Over the last two years, several notable studies have indicated that genome sequencing can make a significant positive impact on newborn health. Bruce Korf, MD, PhD, Director of CCTS Genomic Medicine collaborates on several efforts to increase understanding in this area, with one clinical research study on newborn genome sequencing winding down and another beginning. Both studies explore the benefits of early sequencing and the questions surrounding delivery of results. Here’s the latest update on SouthSeq and BabySeq2.
SouthSeq
Four years ago, SouthSeq was funded by the NIH with the goal to use whole genome sequencing to find the cause of medical problems in newborns in the neonatal intensive care unit (NICU). The SouthSeq protocol also explored the differences, if any, in delivery of results coming from hospital nursery staff instead of genetic counselors. There is a recognized shortage of trained genetic counselors, so training practitioners to share results with patients’ families could increase the ability to deploy genome sequencing on a wider scale. The nursery teams were trained on how to provide families with the information gleaned from testing, and families provided feedback on their satisfaction with the delivery. The sessions were recorded and the audio reviewed by genetic counselors for accuracy, with feedback and corrections given to nursery staff.
The research team was led by investigators at HudsonAlpha and spanned NICUs across Alabama, Kentucky, Louisiana and Mississippi. Participant enrollment has concluded, and results are still being evaluated, but SouthSeq provided a few early observations:
- Sequencing in the nursery often identifies issues that might otherwise be missed and allows for earlier intervention. In many cases, the results of sequencing informed further medical care.
- Nursery and hospital staff have become enthusiastic about genetic testing, and fortunately have continued access to sequencing on a research basis through the Alabama Genomics Health Initiative (AGHI). AGHI is limited, however, in the speed of results, so decisions still need to be made about rapid sequencing as general clinical practice.
- Initial data show that there were very few concerning errors in delivery of results coming from NICU staff versus genetic counseling staff, which could be critical in a landscape where there are not enough genetic counselors to provide test results quickly. As the data from the study are further evaluated we hope to have more information that will allow comparison of outcomes when nursery staff return genomic results as compared with genetic counselors.
- There seems to be a national movement toward rapid whole genome sequencing in newborns as clinicians realize the benefits of early diagnosis, potentially leading to better coverage from health insurance providers.
The Launch of BabySeq2
As SouthSeq winds down, preparation for BabySeq2 is underway. BabySeq2 was funded by NIH in July, spearheaded by investigators at Harvard Medical School and includes many partners, including UAB, and will begin enrolling participants early in 2022. The general difference in BabySeq2 compared with SouthSeq is that recruitment will occur in pediatric practices, among healthy babies ages 1 to 6 months old. The participants will either receive whole genome sequencing or an assessment of family history, with return of certain types of results to parents of the babies. Some of the questions being asked through the study are:
- What kinds of medical information can be obtained from sequencing newborns?
- What results may have a beneficial effect on interventions for the baby? For family members?
- What is the best way to communicate genomic results to parents?
- How do families view the results? Are there concerns of stigmatization? Anxiety over future health events?
Studies like these further national efforts to create more efficiencies in sequencing and delivering results, ultimately providing more access to sequencing and the knowledge it provides. To learn more about either of these studies, contact
This email address is being protected from spambots. You need JavaScript enabled to view it. or theThis email address is being protected from spambots. You need JavaScript enabled to view it. . -
Mining the Genome - Issue 8 / How do we integrate genetic and genomic test results into the context of healthcare?
Historically speaking, Electronic Health Records (EHRs) were not designed to be a landing place for genetic and genomic information, but now that such testing is becoming a prominent part of health care and research, the resulting data must have an effective, reliable presence in EHRs. A team at UAB is addressing this need and Dr. Bruce Korf, director of CCTS Genomic Medicine, shares how.
The EHR Challenge
Genomic medicine involves the use of genomic information to guide medical care, but current electronic health records were not designed to incorporate genomic data. As a result, genomic data, whether obtained through clinical testing or through participation in a research program that returns results to participants, such as AGHI or eMERGE, do not have a standard “home” in the EHR. This could mean a provider may miss the fact that a test has already been conducted by a research team or another provider, and therefore duplicate testing or fail to take account of test results in providing care. Additionally, one genetic testing outfitter may deliver providers results using electronic web portals and others with paper or pdf reports, making the location of results even trickier.
A streamlined process and designated location for results is now becoming a reality at the CCTS Hub, UAB. Within the next year, UAB’s EHR System, IMPACT (Improved Methods of Patient Information Access of Core Clinical Tasks), will include new areas designated for reports specific to genomic sequencing, pharmacogenetic tests, polygenic risk score results and individual single gene or panel- based test results. Ultimately, it is planned to make it possible for providers to order genomic tests directly from IMPACT, and to arrange for pre- and post-test genetic counseling if desired.
Dr. Korf, along with a team that includes Nita Limdi, PharmD, PhD, MSPH, FAHA, Associate Director at the UAB Hugh Kaul Personalized Medicine Institute and Principal Investigator for eMERGE, Renie Moss, Program Director for AGHI and Thalia Baker, FACMPE, Associate Vice President of Primary Care at UAB, are working together to examine both clinical and research workflows to ensure successful implementation of IMPACT’s new addition. As the updates begin to roll out, the team will lead a coordinated effort of outreach and education. The CCTS will continue to provide updates on the progress of this exciting project, but if you have specific questions related to genomic medicine and IMPACT,
This email address is being protected from spambots. You need JavaScript enabled to view it. . -
Mining the Genome - Issue 7/ How Genomic Medicine is Answering Big Questions
This schema helps identify the major questions and specific research projects that aim to address the questions. Use it to identify gaps that need to be filled and current studies that aim to help fill these gaps. The list is undoubtedly incomplete, so please also feel free to add both questions and studies that we may have overlooked.
Opportunities Study Genome Sequencing
- Diagnostic Yield
- Return of Results
- Resolution of VUS
Undiagnosed Diseases Program
SouthSeq
BabySeq
CPAMPopulation Genomics
- Engagement
- Recruitment
- Return of Results
- Outcomes
Alabama Genomic Health Initiative (AGHI)
All of UsPrimary Care Genomics
- Actionable Variants
- Pharmacogenetics
- Polygenic Risk Scores
AGHI
eMERGEEducation
- Students
- Postdocs
- Primary Physicians
SURE-GM
T32 Genomic Medicine
AGHI
BabySeq/SouthSeqBelow are the research questions being addressed by the CCTS genomics projects underway.
Genome Sequencing
Diagnostic Yield:
• How do you decide who to sequence?
• What is the likelihood that a genome sequencing study is going to be helpful to establish a diagnosis?
• What are the technologies available to increase the probability of success in sequencing, and how do you determine what expectations to set for the given individual?
Return of Results:
• How do you communicate with the patient and family once the genomic sequencing results come in?
• How do educate the patient and family about any findings?
• How do you integrate results into the context of care?
• How do you address the implications for family members?
Resolution of a variant of unknown significance (VUS):
• How do you find out if the VUS is causal to the patient’s symptoms?
• How can data sharing help to resolve VUSs?
• What role does creation of model systems play in resolution of VUSs?
The CCTS Genomics Projects that are taking on these questions: Undiagnosed Diseases Program, BabySeq, SouthSeq, CPAM
Population Genomics
Engagement:
• How do you earn people’s trust and address their concerns to make sure they are comfortable with the research process?
• What kinds of concerns are important for people of diverse backgrounds?
Recruitment:
• How do you recruit in a way that makes signing up and participation easy for all people?
Return of Results:
• How do you return the results in a practical and ethically appropriate way?
Outcomes:
• How do you monitor outcomes? What does a person do with the genomic information yielded?
The CCTS Genomics Projects that are taking on these questions: AHGI, All of Us
Primary Care Genomics
Actionable Variants:
• How do you integrate genomics into the point of care for persons with an actionable variant? (Actionable variants are fairly rare, affecting only between 1 and 3 percent of people, but identifying and treating can produce a significant positive impact on their care.)
Pharmacogenetics:
• What information might help adjust a patient’s medication regimen?
• Are there adverse effects that can be avoided if you know in advance that a patient is at risk?
• How do you get medication information to the primary physician and guide them in their use?
Polygenic Risk Scores:
• Can you use genomics to come up with a profile of persopnalized risk of common disease and provide a management scheme to deal with that risk?
The CCTS Genomics Projects that are taking on these questions: AGHI, eMERGE
Education
• How do we train people (at all levels) in genomic medicine?
• How do you integrate genomics with medical care?
The CCTS Genomics Projects that are taking on these questions: SURE-GM, T32 in Genomic Medicine, AGHI, BabySeq/SouthSeq
Interested in any of the projects and/or research questions above?This email address is being protected from spambots. You need JavaScript enabled to view it. to connect with Dr. Korf.
-
Mining the Genome - Issue 6 / UAB Center for Precision Animal Modeling
The UAB Center for Precision Animal Modeling (CPAM) is one of only three centers in the United States funded by the NIH to create animal models for studying disease. The CPAM is still in its first year of its award, and the outreach for projects and collaborators is ramping up. Could the CPAM team and its capabilities have a place in your research?
While the discovery of a genetic variant may lead to an answer for a patient, genetic variants are oftentimes found to be ‘new’, not yet documented in anyone. Though the variant may seem like a plausible candidate to explain the symptoms a patient is experiencing, these variants of unknown significance (VUS) require functional data to provide certainty. Enter precision animal modeling, putting the variants that are discovered in people into various animal model systems to replicate the phenotype in the model. CCTS Genomic Medicine director Dr. Bruce Korf simplifies the concept further: “If we put this variant or mutation in a mouse, does the mouse express symptoms similar to those in the patient?”
While the physiology of animals is obviously different from that of humans, some of the core features can be replicated, providing confidence that the effect of the variant is relevant to mechanisms of disease pathogenesis. From there, potential therapeutics might be administered, making the modeling process a roadmap to treatments. The animal models may therefore be the frontline before clinical trials. One of the first projects the Center began working on was in partnership with the Wolverine Foundation, a nonprofit searching for treatments for the neuro-developmental disease caused by genetic variations in the gene MAPK8IP3. C. elegans (worm) models can be generated very quickly, so the Center got to work and is now testing repurposed FDA-approved drugs hoping to correct the phenotype.
Precise Project Selection
So far, forty proposals have been submitted to the CPAM for modeling consideration, with 12-18 projects moving forward through the pipeline currently. Selections are made by a transdisciplinary team of clinicians, cell biologists, and bioinformaticians, along with CPAM leaders Brad Yoder, Ph.D., chair of the UAB Department of Cell, Developmental and Integrative Biology, and Matt Might, Ph.D., director of the Hugh Kaul Precision Medicine Institute. The committee together determines if animal modeling is warranted for the proposal at hand and if so, which animal(s) should be considered, as each offers specific benefits. “We don’t work as silos like we used to. We lean on expertise from wide breadth of disciplines so that we get feedback from all different angles, and that process is exciting and fun,” shares Dr. Yoder.
Though the selection process is rigorous, Dr. Might highlights the importance of making engagement with the CPAM accessible and valuable, saying, “The CPAM lowers the barrier to entry for clinicians and/or investigators that want to engage in precision disease modeling. In principle, a physician can approach the CPAM with no more than a genetic report and medical history and end up with a model that could either help resolve a diagnostic odyssey or become a platform for further investigation into the disease.”
The CCTS & the CPAM: Pursuing Partnership
There are multiple ways to engage and work with the CPAM. Physicians across the CCTS Partner Network are encouraged to refer patients with chronic, undiagnosed medical conditions for consideration by the Undiagnosed Diseases Program, which serves as a portal to CPAM. Similarly, CCTS investigators can play a vital role in advocating for the priority development of select animal models and/or leverage such models to collaborate in mechanistic studies to help illuminate the biologic impact of genetic variants. Dr. Yoder shares, “We are excited to have models generated in partnership with members of the CCTS, to work together on new models and make them the most useful they can possibly be, but also to utilize the expertise of investigative teams across the Partner Network to thoroughly analyze existing models for further discovery.”
Learn more about the CPAM and submit a request for the generation of an animal model here or contact
This email address is being protected from spambots. You need JavaScript enabled to view it. directly with questions. -
Mining the Genome - Issue 5 / Undiagnosed Diseases Program (UDP)
The Undiagnosed Diseases Program (UDP) is a clinical program that launched at the University of Alabama at Birmingham in 2013. The program is funded by institutional support and revenue generated from patient visits. The UDP aims to discover a diagnosis for patients with rare conditions or unusual presentations of not-so-rare conditions, and in the process, to advance medical knowledge about disease. The UDP team has been successful in assigning a diagnosis to 42% of the patients they’ve evaluated thus far.
The Team
Dr. Bruce Korf, director of CCTS Genomic Medicine and medical geneticist in the UAB Department of Genetics is joined by Dr. Anna C. E. Hurst, medical geneticist and pediatrician in the UAB Department of Genetics and Dr. Martin Rodriquez, internist and infectious disease specialist in the UAB Department of Medicine. The team also includes nurse practitioners Kaitlin Callaway and Tammi Skelton. In addition, physicians from various medical specialties provide their expertise on an as-needed basis.
The Process
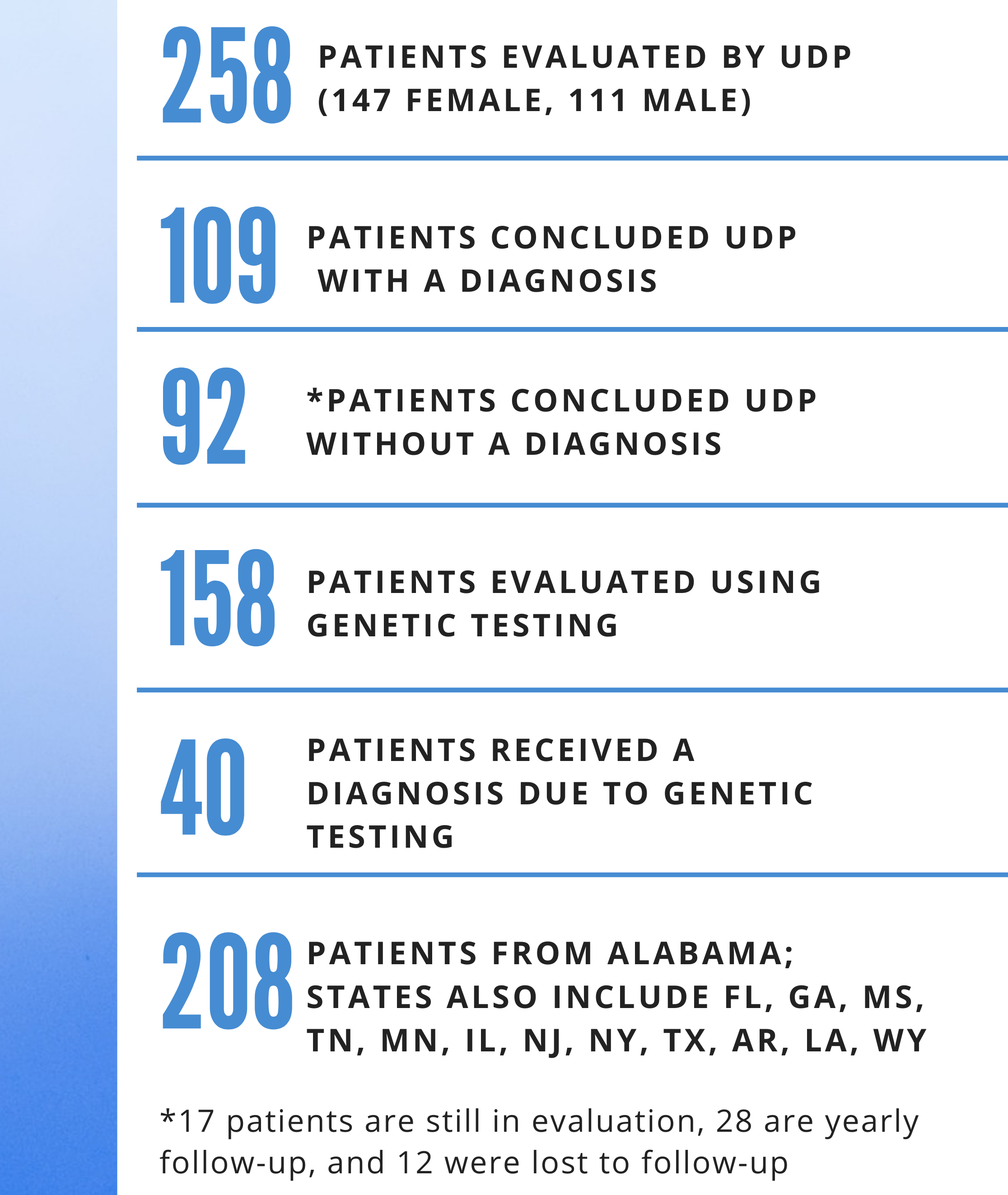 The program operates on referrals from providers across the CCTS Partner Network and beyond with guidelines designed to ensure patients have already exhausted all relevant evaluation options. “We consider each referral carefully, asking ourselves if we can add something that might lead to a diagnosis that up until now has escaped detection,” shares Korf. Patients accepted into the program have been experiencing their symptoms for more than 6 months and have already been extensively evaluated without diagnosis. Once a patient enters the UDP, the first step of the process requires careful assembly of what is usually years, and sometimes decades, of medical records and test results. Nurse practitioners Callaway and Skelton prepare a synopsis of the information, and from there, the entire UDP team gathers to discuss their ideas on what the diagnosis may be. This conversation alone may yield a diagnosis, but oftentimes the team turns to genome sequencing, what Korf calls the single most productive tool at hand. It is also quite convenient for Alabama patients, who are able to receive free sequencing provided by CCTS Partner HudsonAlpha Institute for Biotechnology through the Alabama Genomic Health Initiative. Patients in other states are often able to leverage their health insurance coverage.
The program operates on referrals from providers across the CCTS Partner Network and beyond with guidelines designed to ensure patients have already exhausted all relevant evaluation options. “We consider each referral carefully, asking ourselves if we can add something that might lead to a diagnosis that up until now has escaped detection,” shares Korf. Patients accepted into the program have been experiencing their symptoms for more than 6 months and have already been extensively evaluated without diagnosis. Once a patient enters the UDP, the first step of the process requires careful assembly of what is usually years, and sometimes decades, of medical records and test results. Nurse practitioners Callaway and Skelton prepare a synopsis of the information, and from there, the entire UDP team gathers to discuss their ideas on what the diagnosis may be. This conversation alone may yield a diagnosis, but oftentimes the team turns to genome sequencing, what Korf calls the single most productive tool at hand. It is also quite convenient for Alabama patients, who are able to receive free sequencing provided by CCTS Partner HudsonAlpha Institute for Biotechnology through the Alabama Genomic Health Initiative. Patients in other states are often able to leverage their health insurance coverage.The sequencing often reveals a genetic variant that contains some of the answers patients and their families have been searching for. Then the UDP team looks for more data, often making use of Gene Matcher, an international database that allows providers all over the world to connect and compare notes over their patients’ common symptoms and genetic variants. “Very often you will find that you’ve got a patient that’s very similar to somebody else’s and they share the same or very similar genetic change and that gives you confidence that the genetic change is the underlying cause of the condition,” shares Korf. From there, treatment options can be explored and medical knowledge can be captured. And sometimes, Korf notes, that knowledge is entirely new: “A handful of cases have even resulted in a new discovery, finding a diagnosis that had never been previously described before but turns out to be a new entity that this patient helps to define.” Now, with the UAB Center for Precision Animal Modeling (CPAM), we have the option of generating model systems to further validate new genomic variants and fuel additional discoveries that may be translated to better care of patients who were previously viewed as medical mysteries.
Learn more about the UDP referral process and submit a referral form here. -
Mining the Genome - Issue 4 / Electronic Medical Records and Genomics (eMERGE)
The CCTS sat down with Nita Limdi, Pharm.D, PhD, MSPH, FAHA, professor of neurology and epidemiology, director for the program in Translational Pharmacogenomics, associate director at the UAB Hugh Kaul Personalized Medicine Institute, and principal investigator for eMERGE at UAB, to chat about the plans her team has for the recent eMERGE award they secured last year. Here’s an overview of the goals of eMERGE and what its researchers hope to achieve.
The History of eMERGE
The Electronic Medical Records and Genomics (eMERGE) consortium was launched by the NIH in September of 2007 with the goal of developing, disseminating, and applying approaches to research that combine biorepositories and electronic medical record (EMR) systems for genomic discovery and implementation research. In addition, the consortium reviews ethical issues like privacy and confidentiality and brings together researchers with a wide range of expertise in genomics, statistics, ethics, informatics and clinical medicine.
The Goals of the eMERGE Network
- Calculate validated polygenic risk scores (PRS) for comples diseases, telling you how a person's risk for disease compares to others with a different genetic constitution
- Communicate genomic risk profiles and relevant clinical recommendations based on polygenic risk score (PRS), family history, and other clinical data
- Recruit and genotype 25,000 individuals of diverse ancestry, prospectively calculate their genomic risk for selected conditions and return risk estimates and management recommendations to participants and providers
- Assess uptake of risk-reduction recommendation and impact on related clinical outcomes
Take a deeper look at the aims and goals of the eMERGE Network here.
What Are Genomic Risk Scores?
Every individual carries thousands of genetic variations – typically single letter DNA changes.
For some diseases, (e.g. cystic fibrosis) variation in a single gene (e.g. cystic fibrosis transmembrane conductance regulator; CFTR) confers very high risk of getting a particular disease. However, these monogenic diseases are rare. For most common diseases (e.g. coronary heart disease), variation in many genes each conferring a small risk contributes to the risk of developing disease. Researchers identified these many risk variants (using genome wide association studies) and calculated the overall risk score based on the sum of all the common variants. These polygenic risk scores (PRS), when combined with lifestyle and clinical factors, provide a comprehensive Genome Informed Risk Assessment (GIRA). The PRS can help estimate future risk of common diseases (e.g. heart disease, diabetes, hypertension etc.) and the GIRA can inform treatment interventions (e.g. additional lab tests, medications, lifestyle changes) to tailor preventive strategies for mitigating the disease risk.
UAB Joined eMERGE in 2020 with the Aim of Integrating Genomic Risk Assessment for Chronic Disease Management in a Diverse PopulationIn 2020, Dr. Limdi and James Cimino, MD, were awarded eMERGE funding (making UAB the only site added to the eMERGE consortium this round) that would create even more opportunity for discovery in genomic medicine. Limdi knew that the University of Alabama at Birmingham had diverse cohorts for integration into this study, and she designed the grant to explore those diverse populations. The team’s specific aims are to:
- Select 15 common diseases and race-specific polygenic risk scores (PRS)
- Explore patient perspectives on use of family history and PRS for estimating disease risk among AGHI participants
- Establish methods to implement genomic risk assessment and risk management in clinical care
-
- To recruit 2,500 patients in general medicine practice with greater than 75% being in underserved populations
- To incorporate polygenic risk scores, family history, and clinical data to compute genomic risk estimate for 15 common diseases
- To identify high-risk patients (top 2%) for each disease and deploy risk reduction recommendations
- To assess the uptake of genomic risk assessments, adherence to clinical visits, and clinical outcomes
-
What UAB Brings to the Table
“There have been more than 4,000 genome-wide association studies conducted that have enabled the development of polygenic risk scores, which can be used to predict the risk of many common diseases,” shares Limdi. However, Limdi says those risk scores were derived primarily from patients of European or Asian descent and that patients of African descent are dramatically underrepresented by the scores. “Our challenge will be to recruit and collect more data on underrepresented populations and to use that data to better understand how we can employ genomic scores to predict disease risk in people of other races,” Limdi says.
What may also be most significant about the study is the unique comprehensive approach the team is taking to assessing appropriate clinical care. “The vital first step to leverage the power of genomics to prevent disease is to use genomic risk assessments to identify and — where appropriate — pre-treat at-risk patients,” Limdi says. “At UAB, we will bring our expertise and experience to collaborate with the eMERGE investigative team to take this vital first step."
Limdi looks forward to those results. “We’ve used genomics to diagnose disease. We’ve used genomics to treat disease. But now we are using genomics to see if we can truly prevent disease. And I find that very exciting.” -
Mining the Genome - Issue 3 / Alabama Genomics Health Initiative (AGHI) Will Begin Enrollment with Clinical Practice Partners
The Alabama Genomics Health Initiative (AGHI) is in its fifth year of providing participants genomic testing to identify genetic variants that predict high risk for diseases like cancer and heart disease and genetic counseling to support early interventions and treatments. The partnership between UAB and HudsonAlpha Institute for Biotechnology also includes a focus on research, through which data from test results from across 67 Alabama counties will be used to advance scientific understanding of the role that genes play in health and disease.
Now the AGHI will be embedded into clinical practice in Birmingham, Hoover, and Selma, Alabama, setting the stage for precision care that is better-informed and more cost-effective. CCTS Genomic Medicine Director and AGHI principal investigator Dr. Bruce Korf shares more about the potential of this pilot opportunity.
The Decision to Move to Clinical Practice
In March, the COVID-19 pandemic paused AGHI enrollment, but even in spite of that, AGHI leadership had been contemplating a change in the enrollment strategy. “When we started AGHI, the All of Us Research Program wasn’t active in our region, and even if it had been, it wasn’t offering anything in genomics at the time,” explains Dr. Korf, who leads the southern network of the All of Us Research Program. Now that fact has changed, and All of Us provides participants the same genetic feedback the AGHI does. So the AGHI team asked themselves, “What can we do that All of Us is not able to do, that would preserve our ability to generate a research database while returning value to our participants?” Integrating into medical practices, working hand in hand with providers, was the answer.
Minimizing Disruption in a Clinical Setting
Adding more responsibility to the plates of physicians treating patients while navigating a pandemic is what the AGHI is trying hard to avoid. Renie Moss, AGHI Program Director, works with the physicians who will be launching this pilot, incorporating the AGHI workflow into practice. “The feedback we have received from the physicians is that they are definitely eager to be able to provide information to their patients that would come from AGHI enrollment, but along with that enthusiasm is the request for more information about how this will work in a busy clinic setting,” shares Ms. Moss. The basic workflow sounds simple: physicians enroll their patients, patients consent to the genotyping, and physicians deliver the results, incorporating those results into the patient’s care where appropriate. But the AGHI team is building the systems that will make the process easier with things like:
-
developing an e-consent and telemedicine enrollment process that minimizes interruption of the actual clinical appointment,
-
creating physician educational resources that can be provided online/virtually, both out of respect for busy practitioners and in response to the pandemic, and
-
leaning on established experience of the participant recruitment team in the UAB Recruitment and Retention Shared Facility.
Erin Delaney, MD, the Clinical Medical Director at UAB Highlands, will be among the first to implement the new workflow:
“Our clinic is thrilled to be amongst the first clinics to pilot the integration of AGHI into our practice. We know this will be a learning process for our providers, staff and patients, yet we feel the ultimate benefits, including improved health outcomes, for our community of patients will be long-lasting and certainly worthy of this journey. We are excited about taking the next steps with AGHI to bring this level of medicine to our patients.”
But Wait, There's More
Patients will also have access to pharmacogenetic testing, which provides the genetic underpinnings that can be used to predict their body’s response to certain drugs. Participants who opt-in to pharmacogenetic testing will receive information that could guide which drugs are prescribed to them and at what dosage. Dr. Nita Limdi, Director of the Translational Pharmacogenomics Program and Associate Director of the Hugh Kaul Personalized Medicine Institute, is passionate about the benefit this testing has on both patient health outcomes and financial stewardship. Over $330 billion is spent on medications each year in the United States, and chronic diseases account for 90% of our healthcare expenses. Money is wasted when a drug doesn’t work. Previous trial-and-error approaches in prescribing medications were often met by poor efficacy in large fractions of patients. As Dr. Limdi points out, one might imagine how much time and money could be saved through this pharmacogenetic testing, which can predict treatment response in advance. AGHI will further empower physicians with evidence-based support to make treatment decisions, with guidance from Dr. Limdi and her team. “As we bolster relationships with physicians,” explains Limdi, “this effort will serve as a platform to communicate about the utilization of genetic results in precision medicine and to collect feedback on how we may optimize the use of this technology judiciously.” -
-
Mining the Genome - Issue 2 / All of Us Research Program Update
We’re wrapping up November with our second “Mining the Genome” conversation with Dr. Bruce Korf, CCTS Genomic Medicine Director. Over the course of the month, we’ve introduced our members to the newly launched All of Us Researcher Workbench beta, and in this conversation, Dr. Korf helped provide some quick takeaways for utilizing this resource:
-
UAB is the hub of the All of Us Southern Network, strengthened by the CCTS Partner Network, and was one of the largest enrolling sites in the country before the current pandemic.
-
Reactivation efforts are underway, with emphasis on participant retention, including re-consenting participants to utilize the now-available genome sequencing and genotyping.
-
Researchers from institutions with signed Data Use Agreements can begin using the Researcher Workbench.
-
Researchers can create workspaces within the Workbench to create and analyze cohorts of large numbers of de-identified data sets and access genomic data to look for genomic markers.
-
Enrollment in All of Us is still underway, so the data is not yet complete, but will grow richer as participation grows (All of Us aims to engage a cohort of 1 million or more!)
-
UAB has a signed DUA, so any UAB researcher can access the Workbench beta site at this time.
Click here to learn more and access the recent CCTS training and slide deck on the Researcher Workbench beta. You can also explore additional tutorials here.
-
-
Mining the Genome - Issue 1 / Introduction to Genomic Medicine
An exceptional amount of genomics research activity is underway both locally and nationally, and this column will share new developments and possible implications that will hopefully result in more opportunities for researchers to connect ideas and efforts. Each month, the CCTS will share a conversation with the director of CCTS Genomic Medicine, Bruce Korf, MD, PhD, Wayne H. and Sara Crews Finley Endowed Chair in Medical Genetics, Associate Dean for Genomic Medicine at UAB, and the Chief Genomics Officer for UAB Medicine. Dr. Korf will be providing updates on an array of genomics-related topics and at the end of each column will provide guidance on how you can connect further on that topic.
Let’s kick off this series by establishing a foundation, some genomics groundwork, courtesy of Dr. Korf.
CCTS: What is the overall goal in genomic medicine, from a ‘big picture’ perspective?
Korf: People have been excited about genomics opportunities since the human genome sequence was deciphered, which was now about 17 years ago. But what has become clear is that having the sequence in hand doesn’t automatically translate into better patient care. There’s a lot that has to be done to take that information and develop the systems needed to apply it to patient care, to validate it and demonstrate that it is effective and safe to use. It has therefore taken a fair amount of time to reach this point, where we are now seeing research that is used in routine practice.
From the perspective of an academic medical center, I think there are two things critical for us to keep in mind. We want to provide the best access to cutting edge care that we can—as genomic medicine-related technologies become available and are demonstrated to be effective, we want to make sure our patients have access. But we must also do the work to demonstrate that these approaches have clinical utility. It isn’t just our job to read the book of genomic medicine and apply it to our patients, but to actually write the book.
CCTS: What is genomic medicine?
Korf: Genomics is the study of the entire human genome, the entire complement of genetic material in each of our cells. We have altogether 6 billion base pair of DNA and these encode some 22,000 genes and all of the regulatory processes that turn those genes on or off in the right time and the right place. Genomics really amounts to studying large collections of genetic elements, both genes and control processes to try and put them together into an understanding of how they are important in regulating cell processes, and ultimately contributing to health and disease.
Genetics, in contrast, usually focuses on one gene at a time, and how it is passed from generation to generation and how it functions in the cell, so with genomics we are looking at genes much more globally or collectively. Metaphorically, think of genetics as a book that describes a complicated cellular process and think of genomics as a library of where all of those books are collected.
The concept of genomic medicine is using genomic information to make decisions to either prevent, diagnose, or treat disease. Some of it happens at a at a population level, trying to identify individuals at risk and using that information to try and prevent them from developing disease. Some of it is in achieving a more precise diagnosis in an individual who has symptoms of disease. And finally, genomics also comes into use to identify and guide treatment, and to be sure that treatments being used are used optimally to maximize benefit and minimize side effects.
What initiatives and research developments will Dr. Korf be sharing over the coming months?
Since the genomics landscape is so vast, Korf organizes genomics research into three arenas: rare diseases, common diseases, and cancer, and then conceptualizes each of those into three parts: prevention, diagnosis, and treatment. Korf will use this columns to talk about that entire ‘matrix’ of the genomics world, including work surrounding:
-
The Undiagnosed Diseases Program, which has a 30% success rate in providing a diagnosis for patients who have had great difficulty in receiving a diagnosis
-
The role of genomics in customized cancer therapies
-
The Alabama Genomic Health Initiative, testing Alabamians for the presence of specific genetic variants that may predict the development of serious medical conditions that can be prevented or treated if the diagnosis is made early
-
The All of Us Research Program, an NIH-sponsored initiative to enroll 1 million people to sequence their genomes and collect electronic health record information
-
The eMERGE Consortium, which has a goal to develop polygenic risk scores to identify people at unusually higher risks of disease and use that to institute effective treatments
-
Advancements in Maternal Fetal Medicine preconception counseling and carrier testing
-
The Hugh Kaul Precision Medicine Institute’s work to understand the underlying cause of disease at a genetic level in order to select therapies
-
Developments in pharmacogenetic testing
-
The Center for Precision Animal Models (C-PAM), which is taking genomic variance and developing various animal models that can be used to further study how those gene variants influence disease and treatment.
-